Abstract
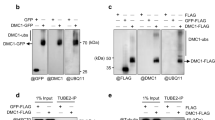
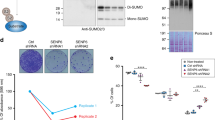
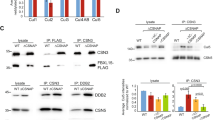
Cdc48 (also known as p97), a conserved chaperone-like ATPase, plays a strategic role in the ubiquitin system1,2,3. Empowered by ATP-driven conformational changes 4, Cdc48 acts as a segregase by dislodging ubiquitylated proteins from their environment1,2,5. Ufd1, a known co-factor of Cdc48, also binds SUMO (ref. 6), but whether SUMOylated proteins are subject to the segregase activity of Cdc48 as well and what these substrates are remains unknown. Here we show that Cdc48 with its co-factor Ufd1 is SUMO-targeted to proteins involved in DNA double-strand break repair. Cdc48 associates with SUMOylated Rad52, a factor that assembles the Rad51 recombinase on chromatin. By acting on the Rad52–Rad51 complex, Cdc48 curbs their physical interaction and displaces the proteins from DNA. Genetically interfering with SUMO-targeting or segregase activity leads to an increase in spontaneous recombination rates, accompanied by aberrant in vivo Rad51 foci formation in yeast and mammalian cells. Our data thus suggest that SUMO-targeted Cdc48 restricts the recombinase Rad51 by counterbalancing the activity of Rad52. We propose that Cdc48, through its ability to associate with co-factors that have affinities for ubiquitin and SUMO, connects the two modification pathways for protein degradation or other regulatory purposes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Jentsch, S. & Rumpf, S. Cdc48 (p97): a molecular gearbox in the ubiquitin pathway? Trends Biochem. Sci. 32, 6–11 (2007).
Stolz, A., Hilt, W., Buchberger, A. & Wolf, D. H. Cdc48: a power machine in protein degradation. Trends Biochem. Sci. 36, 515–523 (2011).
Ghislain, M., Dohmen, R. J., Levy, F. & Varshavsky, A. Cdc48p interacts with Ufd3p, a WD repeat protein required for ubiquitin-mediated proteolysis in Saccharomyces cerevisiae. EMBO J. 15, 4884–4899 (1996).
Rouiller, I. et al. Conformational changes of the multifunction p97 AAA ATPase during its ATPase cycle. Nature Struct. Biol. 9, 950–957 (2002).
Rape, M. et al. Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell 107, 667–677 (2001).
Nie, M. et al. Dual recruitment of Cdc48 (p97)-Ufd1-Npl4 ubiquitin-selective segregase by small ubiquitin-like modifier protein (SUMO) and ubiquitin in SUMO-targeted ubiquitin ligase-mediated genome stability functions. J. Biol. Chem. 287, 29610–29619 (2012).
Ramadan, K. et al. Cdc48/p97 promotes reformation of the nucleus by extracting the kinase Aurora B from chromatin. Nature 450, 1258–1262 (2007).
Raman, M., Havens, C. G., Walter, J. C. & Harper, J. W. A genome-wide screen identifies p97 as an essential regulator of DNA damage-dependent CDT1 destruction. Mol. Cell 44, 72–84 (2011).
Wilcox, A. J. & Laney, J. D. A ubiquitin-selective AAA-ATPase mediates transcriptional switching by remodelling a repressor-promoter DNA complex. Nat. Cell Biol. 11, 1481–1486 (2009).
Acs, K. et al. The AAA-ATPase VCP/p97 promotes 53BP1 recruitment by removing L3MBTL1 from DNA double-strand breaks. Nature Struct. Mol. Biol. 18, 1345–1350 (2011).
Dantuma, N. P. & Hoppe, T. Growing sphere of influence: Cdc48/p97 orchestrates ubiquitin-dependent extraction from chromatin. Trends Cell Biol. 9, 483–491 (2012).
Meerang, M. et al. The ubiquitin-selective segregase VCP/p97 orchestratesthe response to DNA double-strand breaks. Nat. Cell Biol. 13, 1376–1382 (2011).
Mouysset, J. et al. Cell cycle progression requires the CDC-48UFD-1/NPL-4 complex for efficient DNA replication. Proc. Natl Acad. Sci. USA 105, 12879–12884 (2008).
Schuberth, C. & Buchberger, A. Membrane-bound Ubx2 recruits Cdc48 to ubiquitin ligases and their substrates to ensure efficient ER-associated protein degradation. Nat. Cell Biol. 7, 999–1006 (2005).
Neuber, O., Jarosch, E., Volkwein, C., Walter, J. & Sommer, T. Ubx2 links theCdc48 complex to ER-associated protein degradation. Nat. Cell Biol. 7, 993–998 (2005).
Bergink, S. & Jentsch, S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature 458, 461–467 (2009).
Cremona, C. A. et al. Extensive DNA damage-induced sumoylation contributes to replication and repair and acts in addition to the mec1 checkpoint. Mol. Cell 45, 422–432 (2012).
Psakhye, I. & Jentsch, S. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell 151, 807–820 (2012).
Dou, H., Huang, C., Singh, M., Carpenter, P. B. & Yeh, E. T. H. Regulation of DNA repair through deSUMOylation and SUMOylation of replication protein A complex. Mol. Cell 39, 333–345 (2010).
Sacher, M., Pfander, B., Hoege, C. & Jentsch, S. Control of Rad52 recombination activity by double-strand break-induced SUMO modification. Nat. Cell Biol. 8, 1284–1290 (2006).
Shen, Z., Pardington-Purtymun, P. E., Comeaux, J. C., Moyzis, R. K. & Chen, D. J. UBL1, a human ubiquitin-like protein associating with human RAD51/RAD52 proteins. Genomics 36, 271–279 (1996).
Li, W. et al. Regulation of double-strand break-induced mammalian homologous recombination by UBL1, a RAD51-interacting protein. Nucleic Acids Res. 28, 1145–1153 (2000).
Ohuchi, T. et al. Rad52 sumoylation and its involvement in the efficient induction of homologous recombination. DNA Repair. 7, 879–889 (2008).
Krejci, L. et al. Interaction with Rad51 is indispensable for recombination mediator function of Rad52. J. Biol. Chem. 277, 40132–40141 (2002).
Hannich, J. T. et al. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J. Biol. Chem. 280, 4102–4110 (2005).
Perry, J. J., Tainer, J. A. & Boddy, M. N. A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem. Sci. 33, 201–208 (2008).
Song, J., Zhang, Z., Hu, W. & Chen, Y. Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif: a reversal of the bound orientation. J. Biol. Chem. 280, 40122–40129 (2005).
Polo, S. E. & Jackson, S. P. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 25, 409–433 (2011).
Tan, T. L. et al. Mouse Rad54 affects DNA conformation and DNA-damage-induced Rad51 foci formation. Curr. Biol. 9, 325–328 (1999).
Saito, K. et al. The putative nuclear localization signal of the human RAD52 protein is a potential sumoylation site. J. Biochem. 147, 833–842 (2010).
Schermelleh, L. et al. Subdiffraction multicolor imaging of the nuclearperiphery with 3D structured illumination microscopy. Science 320, 1332–1336 (2008).
Yang, H., Li, Q., Fan, J., Holloman, W. K. & Pavletich, N. P. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature 433, 653–657 (2005).
Yuan, S. S. et al. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 59, 3547–3551 (1999).
Jensen, R. B., Carreira, A. & Kowalczykowski, S. C. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature 467, 678–683 (2010).
Moldovan, G. L. et al. Inhibition of homologous recombination by the PCNA-interacting protein PARI. Mol. Cell 45, 75–86 (2012).
Krejci, L. et al. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423, 305–309 (2003).
Rumpf, S. & Jentsch, S. Functional division of substrate processing cofactors of the ubiquitin-selective Cdc48 chaperone. Mol. Cell 21, 261–269 (2006).
Pfander, B., Moldovan, G.L., Sacher, M., Hoege, C. & Jentsch, S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436, 428–433 (2005).
Richly, H. et al. A series of ubiquitin binding factors connects CDC48/p97to substrate multiubiquitylation and proteasomal targeting. Cell 120, 73–84 (2005).
Hoege, C., Pfander, B., Moldovan, G. L., Pyrowolakis, G. & Jentsch, S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419, 135–141 (2002).
Torres-Rosell, J. et al. The Smc5–Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat. Cell Biol. 9, 923–931 (2007).
James, P., Halladay, J. & Craig, E. A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436 (1996).
Acknowledgements
We thank A. Strasser and K. Strasser for technical assistance and I. Psakhye for discussion; J. Gerlach for early contributions; and R. Kanaar (Erasmus MC, NL), S. C. Kowalczykowski (UC Davis, USA) and J. Lukas (CPR, DK) for materials. S.J. is supported by the Max Planck Society and the Louis-Jeantet Foundation; S.J. and S.B. by the RUBICON EU Network of Excellence; M.K. by Boehringer Ingelheim Fonds; S.J. and H.L. by Deutsche Forschungsgemeinschaft SFB646 and Centre for Integrated Protein Science Munich; L.S. and L.H. by Deutsche Forschungsgemeinschaft SFB TR5 and the Bioimaging Network Munich.
Author information
Authors and Affiliations
Contributions
S.B. and S.J. designed the experiments and obtained and analysed the data, T.A. contributed to the mammalian and M.K. the yeast part, L.S. and H.L. planned and conducted the super-resolution microscopy, S.J. and S.B. wrote the paper, and all authors contributed to the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 1362 kb)
Supplementary Table 1
Supplementary Information (XLS 42 kb)
Supplementary Table 2
Supplementary Information (XLS 34 kb)
Supplementary Table 3
Supplementary Information (XLS 28 kb)
Three-dimensional reconstruction of abnormal filamentous Rad51 structures.
Reconstruction of abnormal filamentous Rad51 structures accumulating in U2OS cells 12 h after induction of DNA damage after p97 RNAi treatment. Super-resolution fluorescence microscopy was done with the samples shown in Fig. 5b. The scale represents 1 μm. (MOV 1067 kb)
Rights and permissions
About this article
Cite this article
Bergink, S., Ammon, T., Kern, M. et al. Role of Cdc48/p97 as a SUMO-targeted segregase curbing Rad51–Rad52 interaction. Nat Cell Biol 15, 526–532 (2013). https://doi.org/10.1038/ncb2729
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb2729
This article is cited by
-
Rad51-mediated replication of damaged templates relies on monoSUMOylated DDK kinase
Nature Communications (2022)
-
Nucleolar release of rDNA repeats for repair involves SUMO-mediated untethering by the Cdc48/p97 segregase
Nature Communications (2021)
-
TEX264 coordinates p97- and SPRTN-mediated resolution of topoisomerase 1-DNA adducts
Nature Communications (2020)
-
A SUMO-dependent pathway controls elongating RNA Polymerase II upon UV-induced damage
Scientific Reports (2019)
-
SUMOylation controls stem cell proliferation and regional cell death through Hedgehog signaling in planarians
Cellular and Molecular Life Sciences (2018)