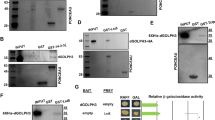
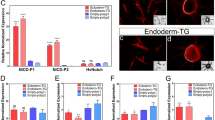
Abstract
The specification of tissue size during development involves the coordinated action of many signalling pathways responding to organ-intrinsic signals, such as morphogen gradients, and systemic cues, such as nutrient status. The conserved Hippo (Hpo) pathway, which promotes both cell-cycle exit and apoptosis, is a major determinant of size control. The pathway core is a kinase cassette, comprising the kinases Hpo and Warts (Wts) and the scaffold proteins Salvador (Sav) and Mats, which inactivates the pro-growth transcriptional co-activator Yorkie (Yki). We performed a split-TEV-based genome-wide RNAi screen for modulators of Hpo signalling. We characterize the Drosophila salt-inducible kinases (Sik2 and Sik3) as negative regulators of Hpo signalling. Activated Sik kinases increase Yki target expression and promote tissue overgrowth through phosphorylation of Sav at Ser 413. As Sik kinases have been implicated in nutrient sensing, this suggests a link between the Hpo pathway and systemic growth control.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Genevet, A. & Tapon, N. The Hippo pathway and apico-basal cell polarity. Biochem. J. 436, 213–224 (2011).
Halder, G. & Johnson, R. L. Hippo signaling: growth control and beyond. Development 138, 9–22 (2011).
Jia, J., Zhang, W., Wang, B., Trinko, R. & Jiang, J. The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev. 17, 2514–2519 (2003).
Pantalacci, S., Tapon, N. & Léopold, P. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat. Cell Biol. 5, 921–927 (2003).
Udan, R. S., Kango-Singh, M., Nolo, R., Tao, C. & Halder, G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat. Cell Biol. 5, 914–920 (2003).
Wu, S., Huang, J., Dong, J. & Pan, D. Hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell 114, 445–456 (2003).
Justice, R. W., Zilian, O., Woods, D. F., Noll, M. & Bryant, P. J. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 9, 534–546 (1995).
Harvey, K. F., Pfleger, C. M. & Hariharan, I. K. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell 114, 457–467 (2003).
Kango-Singh, M. et al. Shar-pei mediates cell proliferation arrest during imaginal disc growth in Drosophila. Development 129, 5719–5730 (2002).
Tapon, N. et al. Salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell 110, 467–478 (2002).
Huang, J., Wu, S., Barrera, J., Matthews, K. & Pan, D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell 122, 421–434 (2005).
Lai, Z-C. et al. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell 120, 675–685 (2005).
Oh, H. & Irvine, K. D. In vivo regulation of Yorkie phosphorylation and localization. Development 135, 1081–1088 (2008).
Dong, J. et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130, 1120–1133 (2007).
Wu, S., Liu, Y., Zheng, Y., Dong, J. & Pan, D. The TEAD/TEF family protein scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev. Cell 14, 388–398 (2008).
Zhang, L. et al. The TEAD/TEF family of transcription factor scalloped mediates hippo signaling in organ size control. Dev. Cell 14, 377–387 (2008).
Goulev, Y. et al. SCALLOPED interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Curr. Biol. 18, 435–441 (2008).
Thompson, B. J. & Cohen, S. M. The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell 126, 767–774 (2006).
Nolo, R., Morrison, C. M., Tao, C., Zhang, X. & Halder, G. The bantammicroRNA is a target of the hippo tumor-suppressor pathway. Curr. Biol. 16, 1895–1904 (2006).
Zhao, B. et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 22, 1962–1971 (2008).
Hao, Y., Chun, A., Cheung, K., Rashidi, B. & Yang, X. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J. Biol. Chem. 283, 5496–5509 (2008).
McCartney, B. M., Kulikauskas, R. M., LaJeunesse, D. R. & Fehon, R. G. The neurofibromatosis-2 homologue, Merlin, and the tumor suppressor expanded function together in Drosophila to regulate cell proliferation and differentiation. Development 127, 1315–1324 (2000).
Hamaratoglu, F. et al. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat. Cell Biol. 8, 27–36 (2006).
Maitra, S., Kulikauskas, R. M., Gavilan, H. & Fehon, R. G. The tumor suppressors Merlin and Expanded function cooperatively to modulate receptor endocytosis and signaling. Curr. Biol. 16, 702–709 (2006).
Poon, C. L. C., Lin, J. I., Zhang, X. & Harvey, K. F. The sterile 20-like kinase Tao-1 controls tissue growth by regulating the Salvador–Warts–Hippo pathway. Dev. Cell 21, 896–906 (2011).
Boggiano, J. C., Vanderzalm, P. J. & Fehon, R. G. Tao-1 phosphorylates Hippo/MST kinases to regulate the Hippo–Salvador–Warts tumor suppressor pathway. Dev. Cell 21, 888–895 (2011).
Baumgartner, R., Poernbacher, I., Buser, N., Hafen, E. & Stocker, H. The WWdomain protein Kibra acts upstream of Hippo in Drosophila. Dev. Cell 18, 309–316 (2010).
Genevet, A., Wehr, M. C., Brain, R., Thompson, B. J. & Tapon, N. Kibra is aregulator of the Salvador/Warts/Hippo signaling network. Dev. Cell 18, 300–308 (2010).
Yu, J. et al. Kibra functions as a tumor suppressor protein that regulatesHippo signaling in conjunction with Merlin and Expanded. Dev. Cell 18, 288–299 (2010).
Badouel, C. et al. The FERM-domain protein expanded regulates Hippo pathway activity via direct interactions with the transcriptional activator Yorkie. Dev. Cell 16, 411–420 (2009).
Oh, H., Reddy, B. V. V. G. & Irvine, K. D. Phosphorylation-independent repression of Yorkie in Fat-Hippo signaling. Dev. Biol. 335, 188–197 (2009).
Zhao, B. et al. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 25, 51–63 (2011).
Paramasivam, M., Sarkeshik, A., Yates, J. R. 3rd, Fernandes, M. J. G. & McCollum, D. Angiomotin family proteins are novel activators of the LATS2 kinase tumor suppressor. Mol. Biol. Cell 22, 3725–3733 (2011).
Chan, S. W. et al. Hippo pathway-independent restriction of TAZ and YAP by angiomotin. J. Biol. Chem. 286, 7018–7026 (2011).
Wang, W., Huang, J. & Chen, J. Angiomotin-like proteins associate with and negatively regulate YAP1. J. Biol. Chem. 286, 4364–4370 (2011).
Bennett, F. C. & Harvey, K. F. Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr. Biol. 16, 2101–2110 (2006).
Cho, E. et al. Delineation of a Fat tumor suppressor pathway. Nat. Genet. 38, 1142–1150 (2006).
Mao, Y. et al. Dachs: an unconventional myosin that functions downstream of Fat to regulate growth, affinity and gene expression in Drosophila. Development 133, 2539–2551 (2006).
Silva, E., Tsatskis, Y., Gardano, L., Tapon, N. & McNeill, H. The tumor-suppressor gene fat controls tissue growth upstream of expanded in the hippo signaling pathway. Curr. Biol. 16, 2081–2089 (2006).
Willecke, M. et al. The fat cadherin acts through the hippo tumor-suppressor pathway to regulate tissue size. Curr. Biol. 16, 2090–2100 (2006).
Chen, C-L. et al. The apical-basal cell polarity determinant Crumbs regulates Hippo signaling in Drosophila. Proc. Natl Acad. Sci. USA 107, 15810–15815 (2010).
Ling, C. et al. The apical transmembrane protein Crumbs functions as a tumor suppressor that regulates Hippo signaling by binding to Expanded. Proc. Natl Acad. Sci. USA 107, 10532–10537 (2010).
Robinson, B. S., Huang, J., Hong, Y. & Moberg, K. H. Crumbs regulates Salvador/Warts/Hippo signaling in Drosophila via the FERM-domain protein Expanded. Curr. Biol. 20, 582–590 (2010).
Varelas, X. et al. The Hippo pathway regulates Wnt/β-catenin signaling. Dev. Cell 18, 579–591 (2010).
Zhao, B., Li, L., Tumaneng, K., Wang, C-Y. & Guan, K-L. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(β-TRCP). Genes Dev. 24, 72–85 (2010).
Schlegelmilch, K. et al. Yap1 acts downstream of α-catenin to control epidermal proliferation. Cell 144, 782–795 (2011).
Silvis, M. R. et al. α-catenin is a tumor suppressor that controls cell accumulation by regulating the localization and activity of the transcriptional coactivator Yap1. Sci. Signal. 4, 1–11 (2011).
Yi, C. et al. A tight junction-associated Merlin-angiomotin complex mediates Merlin’s regulation of mitogenic signaling and tumor suppressive functions. Cancer Cell 19, 527–540 (2011).
Rogulja, D., Rauskolb, C. & Irvine, K. D. Morphogen control of wing growth through the Fat signaling pathway. Dev. Cell 15, 309–321 (2008).
Densham, R. M. et al. MST kinases monitor actin cytoskeletal integrity and signal via c-Jun N-terminal kinase stress-activated kinase to regulate p21Waf1/Cip1 stability. Mol. Cell. Biol. 29, 6380–6390 (2009).
Dupont, S. et al. Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183 (2011).
Fernández, B. G. et al. Actin-Capping Protein and the Hippo pathway regulate F-actin and tissue growth in Drosophila. Development 138, 2337–2346 (2011).
Sansores-Garcia, L. et al. Modulating F-actin organization induces organ growth by affecting the Hippo pathway. EMBO J. 30, 2325–2335 (2011).
Wada, K-I., Itoga, K., Okano, T., Yonemura, S. & Sasaki, H. Hippo pathway regulation by cell morphology and stress fibers. Development 138, 3907–3914 (2011).
Wehr, M. C. et al. Monitoring regulated protein-protein interactions using split TEV. Nat. Methods 3, 985–993 (2006).
Wehr, M. C., Reinecke, L., Botvinnik, A. & Rossner, M. J. Analysis of transient phosphorylation-dependent protein-protein interactions in living mammalian cells using split-TEV. BMC Biotechnol. 8, 55 (2008).
Ribeiro, P. S. et al. Combined functional genomic and proteomic approaches identify a PP2A complex as a negative regulator of Hippo signaling. Mol. Cell 39, 521–534 (2010).
Praskova, M., Khoklatchev, A., Ortiz-Vega, S. & Avruch, J. Regulation of the MST1 kinase by autophosphorylation, by the growth inhibitory proteins, RASSF1 and NORE1, and by Ras. Biochem. J. 381, 453–462 (2004).
Dentin, R. et al. Insulin modulates gluconeogenesis by inhibition of the coactivator TORC2. Nature 449, 366–369 (2007).
Screaton, R. A. et al. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell 119, 61–74 (2004).
Horike, N. et al. Adipose-specific expression, phosphorylation of Ser794 in insulin receptor substrate-1, and activation in diabetic animals of salt-inducible kinase-2. J. Biol. Chem. 278, 18440–18447 (2003).
Okamoto, M., Takemori, H. & Katoh, Y. Salt-inducible kinase in steroidogenesis and adipogenesis. Trends Endocrinol. Metab. 15, 21–26 (2004).
Choi, S., Kim, W. & Chung, J. Drosophila salt-inducible kinase (SIK) regulates starvation resistance through cAMP-response element-binding protein (CREB)-regulated transcription coactivator (CRTC). J. Biol. Chem. 286, 2658–2664 (2011).
Lizcano, J. M. et al. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 23, 833–843 (2004).
Wang, B. et al. A hormone-dependent module regulating energy balance. Cell 145, 596–606 (2011).
Shackelford, D. B. & Shaw, R. J. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat. Rev. Cancer 9, 563–575 (2009).
Nguyen, H. B., Babcock, J. T., Wells, C. D. & Quilliam, L. A. LKB1 tumor suppressor regulates AMP kinase/mTOR-independent cell growth and proliferation via the phosphorylation of Yap. Oncogenehttp://dx.doi.org/10.1038/onc.2012.431 (2012).
Mihaylova, M. M. et al. Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell 145, 607–621 (2011).
Wang, B. et al. The insulin-regulated CREB coactivator TORC promotes stress resistance in Drosophila. Cell Metab. 7, 434–444 (2008).
Straßburger, K., Tiebe, M., Pinna, F., Breuhahn, K. & Teleman, A. A. Insulin/IGF signaling drives cell proliferation in part via Yorkie/YAP. Dev. Biol. 367, 187–196 (2012).
Yu, F-X. et al. Regulation of the Hippo–YAP pathway by G-protein-coupled receptor signaling. Cell 150, 780–791 (2012).
Ahmed, A. A. et al. SIK2 is a centrosome kinase required for bipolar mitotic spindle formation that provides a potential target for therapy in ovarian cancer. Cancer Cell 18, 109–121 (2010).
Charoenfuprasert, S. et al. Identification of salt-inducible kinase 3 as a novel tumor antigen associated with tumorigenesis of ovarian cancer. Oncogene 30, 3570–3584 (2011).
Imielinski, M. et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 150, 1107–1120 (2012).
Hall, C. A. et al. Hippo pathway effector Yap is an ovarian cancer oncogene. Cancer Res. 70, 8517–8525 (2010).
Zhang, X. et al. The Hippo pathway transcriptional co-activator, YAP, is an ovarian cancer oncogene. Oncogene 30, 2810–2822 (2011).
Overholtzer, M. et al. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc. Natl Acad. Sci. USA 103, 12405–12410 (2006).
Kapust, R. B. et al. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 14, 993–1000 (2001).
Bischof, J., Maeda, R. K., Hediger, M., Karch, F. & Basler, K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl Acad. Sci. USA 104, 3312–3317 (2007).
Colombani, J., Polesello, C., Josué, F. & Tapon, N. Dmp53 activates the Hippo pathway to promote cell death in response to DNA damage. Curr. Biol. 16, 1453–1458 (2006).
Acknowledgements
We thank A. Kerai for help with re-screening candidates; T. Gilbank, S. Maloney and F. Earl for maintaining flies; L. Muthusamy for sequencing and helping with plating RNAi libraries; the Vienna Drosophila RNAi Centre for fly stocks; and O. Shaham and B. Thompson for comments on the manuscript. We are very grateful to P. Gaspar for help with fly genetics; F. Josue and P. Langton for advice; and A. Teleman and A. Celik for sharing reagents and exchanging unpublished data before publication. We thank K-A. Nave (Göttingen) for ongoing support. M.C.W. and M.J.R. acknowledge the support by the Bundesministerium für Wirtschaft und Forschung (grants 03EFT6N1 and 16V0008). M.C.W. was supported by an EMBO Long-Term Fellowship. The Tapon laboratory is supported by Cancer Research UK.
Author information
Authors and Affiliations
Contributions
M.C.W. designed the study and experiments and performed most of the experiments. M.V.H. cloned and analysed the full-length Sik3 ORFs and performed the experiments in Figs 5f–i and 8g–j and Supplementary Figs S3k,l and S4f–l. I.G. performed the experiments in Fig. 5a–e and Supplementary Fig. S4a–e and contributed western blot data. T.M.M. contributed phosphatase-treated western blot data, and E.C. helped with cloning. R.E.S., M.J., R.I. and M.H. helped perform and analyse the screen. M.J.R. provided the split TEV technology and supported the study. N.T. conceived and supervised the study. M.C.W. and N.T. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 1267 kb)
Supplementary Table 1
Supplementary Information (XLSX 21 kb)
Supplementary Table 2
Supplementary Information (PDF 61 kb)
Rights and permissions
About this article
Cite this article
Wehr, M., Holder, M., Gailite, I. et al. Salt-inducible kinases regulate growth through the Hippo signalling pathway in Drosophila. Nat Cell Biol 15, 61–71 (2013). https://doi.org/10.1038/ncb2658
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb2658
This article is cited by
-
The LKB1–TSSK1B axis controls YAP phosphorylation to regulate the Hippo–YAP pathway
Cell Death & Disease (2024)
-
SHANK2 is a frequently amplified oncogene with evolutionarily conserved roles in regulating Hippo signaling
Protein & Cell (2021)
-
The potent roles of salt-inducible kinases (SIKs) in metabolic homeostasis and tumorigenesis
Signal Transduction and Targeted Therapy (2020)
-
Mutant GNAS drives pancreatic tumourigenesis by inducing PKA-mediated SIK suppression and reprogramming lipid metabolism
Nature Cell Biology (2018)
-
A feed forward loop enforces YAP/TAZ signaling during tumorigenesis
Nature Communications (2018)