Abstract
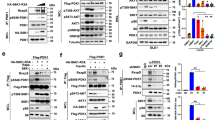
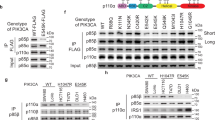
Pyruvate kinase M2 (PKM2) is upregulated in multiple cancer types and contributes to the Warburg effect by unclear mechanisms. Here we demonstrate that EGFR-activated ERK2 binds directly to PKM2 Ile 429/Leu 431 through the ERK2 docking groove and phosphorylates PKM2 at Ser 37, but does not phosphorylate PKM1. Phosphorylated PKM2 Ser 37 recruits PIN1 for cis–trans isomerization of PKM2, which promotes PKM2 binding to importin α5 and translocating to the nucleus. Nuclear PKM2 acts as a coactivator of β-catenin to induce c-Myc expression, resulting in the upregulation of GLUT1, LDHA and, in a positive feedback loop, PTB-dependent PKM2 expression. Replacement of wild-type PKM2 with a nuclear translocation-deficient mutant (S37A) blocks the EGFR-promoted Warburg effect and brain tumour development in mice. In addition, levels of PKM2 Ser 37 phosphorylation correlate with EGFR and ERK1/2 activity in human glioblastoma specimens. Our findings highlight the importance of nuclear functions of PKM2 in the Warburg effect and tumorigenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Change history
27 June 2014
In the online version of this Article originally published, the company name 'Signalway Biotechnology' should have read 'Signalway Antibody'. This has now been corrected in all online versions of the Article.
References
Vander Heiden, M. G., Cantley, L. C. & Thompson, C. B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009).
Koppenol, W. H., Bounds, P. L. & Dang, C. V. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat. Rev. Cancer 11, 325–337.
Cairns, R. A., Harris, I. S. & Mak, T. W. Regulation of cancer cell metabolism. Nat. Rev. Cancer 11, 85–95.
Altenberg, B. & Greulich, K. O. Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics 84, 1014–1020 (2004).
Majumder, P. K. et al. mTOR inhibition reverses Akt-dependent prostate intraepithelial neoplasia through regulation of apoptotic and HIF-1-dependent pathways. Nature Med. 10, 594–601 (2004).
David, C. J., Chen, M., Assanah, M., Canoll, P. & Manley, J. L. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature 463, 364–368.
Clower, C. V. et al. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc. Natl Acad. Sci. USA 107, 1894–1899.
Christofk, H. R. et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 452, 230–233 (2008).
Vander Heiden, M. G. et al. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science 329, 1492–1499.
Yang, W. et al. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature 480, 118–122 (2011).
Luo, W. et al. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell 145, 732–744 (2011).
Gao, X., Wang, H., Yang, J. J., Liu, X. & Liu, Z. R. Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol. Cell 45, 598–609 (2012).
Lu, Z. Nonmetabolic functions of pyruvate kinase isoform M2 in controlling cell cycle progression and tumorigenesis. Chin. J. Cancer 31, 5–7 (2012).
Yang, W. et al. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell 150, 685–696 (2012).
Lu, Z. & Xu, S. ERK1/2 MAP kinases in cell survival and apoptosis. IUBMB Life 58, 621–631 (2006).
Lu, K. P. & Zhou, X. Z. The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease. Nat. Rev. Mol. Cell Biol. 8, 904–916 (2007).
Zheng, Y. et al. FAK phosphorylation by ERK primes ras-induced tyrosine dephosphorylation of FAK mediated by PIN1 and PTP-PEST. Mol. Cell 35, 11–25 (2009).
Lufei, C. & Cao, X. Nuclear import of Pin1 is mediated by a novel sequence in the PPIase domain. FEBS Lett. 583, 271–276 (2009).
Jans, D. A., Xiao, C. Y. & Lam, M. H. Nuclear targeting signal recognition: a key control point in nuclear transport? Bioessays 22, 532–544 (2000).
Mason, D. A., Stage, D. E. & Goldfarb, D. S. Evolution of the metazoan-specific importin alpha gene family. J. Mol. Evol. 68, 351–365 (2009).
Lu, Z., Ghosh, S., Wang, Z. & Hunter, T. Downregulation of caveolin-1 function by EGF leads to the loss of E-cadherin, increased transcriptional activity of beta-catenin, and enhanced tumor cell invasion. Cancer Cell 4, 499–515 (2003).
DeBerardinis, R. J., Lum, J. J., Hatzivassiliou, G. & Thompson, C. B. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 7, 11–20 (2008).
Dang, C. V., Kim, J. W., Gao, P. & Yustein, J. The interplay between MYC and HIF in cancer. Nat. Rev. Cancer 8, 51–56 (2008).
Warburg, O. On the origin of cancer cells. Science 123, 309–314 (1956).
Hsu, P. P. & Sabatini, D. M. Cancer cell metabolism: Warburg and beyond. Cell 134, 703–707 (2008).
Lu, Z. et al. Activation of protein kinase C triggers its ubiquitination and degradation. Mol. Cell Biol. 18, 839–845 (1998).
Xia, Y. et al. c-Jun downregulation by HDAC3-dependent transcriptional repression promotes osmotic stress-induced cell apoptosis. Mol. Cell 25, 219–232 (2007).
Fang, D. et al. Phosphorylation of β-catenin by AKT promotes beta-catenin transcriptional activity. J. Biol. Chem. 282, 11221–11229 (2007).
Acknowledgements
This work was supported by National Institute of Health grants 2R01CA109035 (Z.L.), R01GM068566 (X.C.), R01GM56302 (L.C.C.) and CA16672 (Cancer Center Support Grant) and a research grant (RP110252; Z.L.) from the Cancer Prevention and Research Institute of Texas (CPRIT).
Author information
Authors and Affiliations
Contributions
This study was conceived by Z.L.; Z.L. and W.Y. designed the study; W.Y., Y.Z., Y.X., H.J. and C.A.L. carried out experiments; K.A. provided pathology assistance; X.C., F.G. and L.C.C. provided reagents and conceptual advice; Z.L. wrote the paper with comments from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 1267 kb)
Rights and permissions
About this article
Cite this article
Yang, W., Zheng, Y., Xia, Y. et al. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat Cell Biol 14, 1295–1304 (2012). https://doi.org/10.1038/ncb2629
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb2629
This article is cited by
-
Mechanosensitive channel of large conductance enhances the mechanical stretching-induced upregulation of glycolysis and oxidative metabolism in Schwann cells
Cell Communication and Signaling (2024)
-
Transcriptional regulation and post-translational modifications in the glycolytic pathway for targeted cancer therapy
Acta Pharmacologica Sinica (2024)
-
Stabilization of Pin1 by USP34 promotes Ubc9 isomerization and protein sumoylation in glioma stem cells
Nature Communications (2024)
-
TREM2 deficiency impairs the energy metabolism of Schwann cells and exacerbates peripheral neurological deficits
Cell Death & Disease (2024)
-
A novel protein extracted from Hemerocallis citrina Borani inhibits hepatocellular carcinoma cell proliferation by regulating mitochondria-dependent apoptosis and aerobic glycolysis
Food Science and Biotechnology (2024)