Abstract
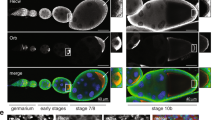
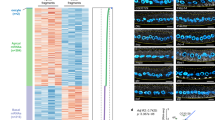
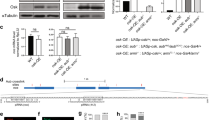
The primary embryonic axes in flies, frogs and fish are formed through translational regulation of localized transcripts before fertilization1. In Drosophila melanogaster, the axes are established through the transport and translational regulation of gurken (grk) and bicoid (bcd) messenger RNA in the oocyte and embryo1. Both transcripts are translationally silent while being localized within the oocyte along microtubules by cytoplasmic dynein1,2,3,4. Once localized, grk is translated at the dorsoanterior of the oocyte to send a TGF- α signal to the overlying somatic cells5. In contrast, bcd is translationally repressed in the oocyte until its activation in early embryos when it forms an anteroposterior morphogenetic gradient6. How this differential translational regulation is achieved is not fully understood. Here, we address this question using ultrastructural analysis, super-resolution microscopy and live-cell imaging. We show that grk and bcd ribonucleoprotein (RNP) complexes associate with electron-dense bodies that lack ribosomes and contain translational repressors. These properties are characteristic of processing bodies (P bodies), which are considered to be regions of cytoplasm where decisions are made on the translation and degradation of mRNA. Endogenous grk mRNA forms dynamic RNP particles that become docked and translated at the periphery of P bodies, where we show that the translational activator Oo18 RNA-binding protein (Orb, a homologue of CEPB) and the anchoring factor Squid (Sqd) are also enriched. In contrast, an excess of grk mRNA becomes localized inside the P bodies, where endogenous bcd mRNA is localized and translationally repressed. Interestingly, bcd mRNA dissociates from P bodies in embryos following egg activation, when it is known to become translationally active. We propose a general principle of translational regulation during axis specification involving remodelling of transport RNPs and dynamic partitioning of different transcripts between the translationally active edge of P bodies and their silent core.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
St Johnston, D. Moving messages: the intracellular localization of mRNAs. Nat. Rev. Mol. Cell Biol. 6, 363–375 (2005).
Weil, T. T., Forrest, K. M. & Gavis, E. R. Localization of bicoid mRNA in late oocytes is maintained by continual active transport. Dev. Cell 11, 251–262 (2006).
Cha, B. J., Koppetsch, B. S. & Theurkauf, W. E. In vivo analysis of Drosophila bicoid mRNA localization reveals a novel microtubule-dependent axis specification pathway. Cell 106, 35–46 (2001).
Clark, A., Meignin, C. & Davis, I. A Dynein-dependent shortcut rapidly delivers axis determination transcripts into the Drosophila oocyte. Development 134, 1955–1965 (2007).
Neuman-Silberberg, F. S. & Schüpbach, T. The Drosophila dorsoventral patterning gene gurken produces a dorsally localized RNA and encodes a TGFa-like protein. Cell 75, 165–174 (1993).
St Johnston, D., Driever, W., Berleth, T., Richstein, S. & Nüsslein-Volhard, C. Multiple steps in the localization of bicoid RNA to the anterior pole of the Drosophila oocyte. Development 107 (Suppl.), 13–19 (1989).
Delanoue, R., Herpers, B., Soetaert, J., Davis, I. & Rabouille, C. Drosophila Squid/hnRNP helps Dynein switch from a gurken mRNA transport motor to an ultrastructural static anchor in sponge bodies. Dev. Cell 13, 523–538 (2007).
Wilsch-Brauninger, M., Schwarz, H. & Nusslein-Volhard, C. A sponge-like structure involved in the association and transport of maternal products during Drosophila oogenesis. J. Cell Biol. 139, 817–829 (1997).
Snee, M. J. & Macdonald, P. M. Dynamic organization and plasticity of sponge bodies. Dev. Dyn. 238, 918–930 (2009).
Beckham, C. et al. The DEAD-box RNA helicase Ded1p affects and accumulates in Saccharomyces cerevisiae P-bodies. Mol. Biol. Cell 19, 984–993 (2008).
Minshall, N., Kress, M., Weil, D. & Standart, N. Role of p54 RNA helicase activity and its C-terminal domain in translational repression, P-body localization and assembly. Mol. Biol. Cell 20, 2464–2472 (2009).
Herpers, B. & Rabouille, C. mRNA localization and ER-based protein sorting mechanisms dictate the use of transitional endoplasmic reticulum–Golgi units involved in gurken transport in Drosophila oocytes. Mol. Biol. Cell 15, 5306–5317 (2004).
Nakamura, A., Amikura, R., Hanyu, K. & Kobayashi, S. Me31B silences translation of oocyte-localizing RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis. Development 128, 3233–3242 (2001).
Dobbie, I. et al. in Live Cell Imaging: A Laboratory Manual Second edn (eds Goldman, Robert D., Swedlow, Jason R. & Spector, David L.) 203–214 (Cold Spring Harbor Laboratory Press, 2010).
Jaramillo, A. M., Weil, T. T., Goodhouse, J., Gavis, E. R. & Schüpbach, T. The dynamics of fluorescently labeled endogenous gurken mRNA in Drosophila. J. Cell Sci. 121, 887–894 (2008).
Forrest, K. M. & Gavis, E. R. Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr. Biol. 13, 1159–1168 (2003).
Bertrand, E. et al. Localization of ASH1 mRNA particles in living yeast. Mol. Cell 2, 437–445 (1998).
Teixeira, D., Sheth, U., Valencia-Sanchez, M. A., Brengues, M. & Parker, R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11, 371–382 (2005).
Chang, J. S., Tan, L., Wolf, M. R. & Schedl, P. Functioning of the Drosophila orb gene in gurken mRNA localization and translation. Development 128, 3169–3177 (2001).
Neuman-Silberberg, F. S. & Schüpbach, T. Dorsoventral axis formation in Drosophila depends on the correct dosage of the gene gurken. Development 120, 2457–2463 (1994).
Wilkie, G. S. & Davis, I. Drosophila wingless and pair-rule transcripts localize apically by dynein-mediated transport of RNA particles. Cell 105, 209–219 (2001).
MacDougall, N., Clark, A., MacDougall, E. & Davis, I. Drosophila gurken (TGFα) mRNA localizes as particles that move within the oocyte in two dynein-dependent steps. Dev. Cell 4, 307–319 (2003).
Bokel, C., Dass, S., Wilsch-Brauninger, M. & Roth, S. Drosophila Cornichon acts as cargo receptor for ER export of the TGFα-like growth factor Gurken. Development 133, 459–470 (2006).
Weil, T. T., Parton, R., Davis, I. & Gavis, E. R. Changes in bicoid mRNA anchoring highlight conserved mechanisms during the oocyte-to-embryo transition. Curr. Biol. 18, 1055–1061 (2008).
Horner, V. L. et al. The Drosophila calcipressin sarah is required for several aspects of egg activation. Curr. Biol. 16, 1441–1446 (2006).
Moser, J. J. & Fritzler, M. J. Cytoplasmic ribonucleoprotein (RNP) bodies and their relationship to GW/P bodies. Int. J Biochem. Cell Biol. 42, 828–843 (2010).
Buchan, J. R. & Parker, R. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell 36, 932–941 (2009).
King, M. L., Messitt, T. J. & Mowry, K. L. Putting RNAs in the right place at the right time: RNA localization in the frog oocyte. Biol. Cell 97, 19–33 (2005).
Kloc, M., Bilinski, S. & Etkin, L. D. The Balbiani body and germ cell determinants: 150 years later. Curr. Top Dev. Biol. 59, 1–36 (2004).
Updike, D. & Strome, S. P granule assembly and function in Caenorhabditis elegans germ cells. J. Androl. 31, 53–60 (2010).
Brangwynne, C. P. et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732 (2009).
Eulalio, A., Behm-Ansmant, I., Schweizer, D. & Izaurralde, E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell Biol. 27, 3970–3981 (2007).
Parker, R. & Sheth, U. P bodies and the control of mRNA translation and degradation. Mol. Cell 25, 635–646 (2007).
Balagopal, V. & Parker, R. Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr. Opin. Cell Biol. 21, 403–408 (2009).
Stoecklin, G., Mayo, T. & Anderson, P. ARE-mRNA degradation requires the 5′- 3′ decay pathway. EMBO Rep. 7, 72–77 (2006).
Buszczak, M. et al. The carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics 175, 1505–1531 (2007).
Theurkauf, W. E. & Hazelrigg, T. I. In vivo analyses of cytoplasmic transport and cytoskeletal organization during Drosophila oogenesis: characterization of a multi-step anterior localization pathway. Development 125, 3655–3666 (1998).
Norvell, A., Debec, A., Finch, D., Gibson, L. & Thoma, B. Squid is required for efficient posterior localization of oskar mRNA during Drosophila oogenesis. Dev. Genes Evol. 215, 340–349 (2005).
Herpers, B., Xanthakis, D. & Rabouille, C. ISH–IEM: a sensitive method to detect endogenous mRNAs at the ultrastructural level. Nat. Protoc. 5, 678–687 (2010).
Slot, J. W. & Geuze, H. J. Cryosectioning and immunolabeling. Nat. Protoc. 2, 2480–2491 (2007).
Rabouille, C. Quantitative aspects of immunogold labeling in embedded and nonembedded sections. Methods Mol Biol. 117, 125–144 (1999).
Filardo, P. & Ephrussi, A. Bruno regulates gurken during Drosophila oogenesis. Mech. Dev. 120, 289–297 (2003).
Hammond, L. E., Rudner, D. Z., Kanaar, R. & Rio, D. C. Mutations in the hrp48 gene, which encodes a Drosophila heterogeneous nuclear ribonucleoprotein particle protein, cause lethality and developmental defects and affect P-element third-intron splicing in vivo. Mol. Cell Biol. 17, 7260–7267 (1997).
Rehwinkel, J., Behm-Ansmant, I., Gatfield, D. & Izaurralde, E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA 11, 1640–1647 (2005).
Court, F. A., Hendriks, W. T., MacGillavry, H. D., Alvarez, J. & van Minnen, J. Schwann cell to axon transfer of ribosomes: toward a novel understanding of the role of glia in the nervous system. J. Neurosci. 28, 11024–11029 (2008).
Parton, R. M., Valles, A. M., Dobbie, I. M. & Davis, I. Live cell imaging in Drosophila melanogaster. Cold Spring Harb. Protoc. 2010, 387–418 (2010).
Weil, T. T., Parton, R. M. & Davis, I. Preparing individual Drosophila egg chambers for live imaging. J. Vis. Exp.http://dx.doi.org/10.3791/3679 (2012).
Gross, D. J. & Webb, W. W. Spectroscopic Membrane Probe, Vol. II. 19–48 (CRC Press Inc., 1988).
Saxton, M. J. & Jacobson, K. Single-particle tracking: applications to membrane dynamics. Annu. Rev. Biophys. Biomol. Struct. 26, 373–399 (1997).
Tadakuma, H., Ishihama, Y., Shibuya, T., Tani, T. & Funatsu, T. Imaging of single mRNA molecules moving within a living cell nucleus. Biochem. Biophys. Res. Commun. 344, 772–779 (2006).
Acknowledgements
We are grateful to: E. R. Gavis (Princeton University, USA), D. Ish-Horowicz (CRUK, UK), T. Schüpbach (Princeton University, USA), A. Jaramillo (Princeton University, USA), J. van Minnen (University of Calgary, Canada), L. Cooley (Yale University, USA), R. Singer (Albert Einstein College of Medicine, USA) and A. Nakamura (RIKEN, Japan) for fly stocks, antibodies and reagents; all members of the Cell Microscopy Center in Utrecht, Netherlands for assistance with electron microscopy; J. W. Sedat (UCSF, USA) and D. Agard (UCSF, USA) for advise on super-resolution imaging; E. R. Gavis for experimental advice; A. Ephrussi (EMBL, Germany) for discussions of the data. This work was supported by: Marie Curie International Incoming Fellowship (ROXA0) to T.T.W.; Wellcome Trust Senior Research Fellowship (081858) to I.D. and supporting R.M.P. and T.T.W.; Cancer Research UK funding to R.H.; studentship from Wellcome Trust to J.M.H.; Darwin Trust to J.S.
Author information
Authors and Affiliations
Contributions
T.T.W., R.M.P., C.R. and I.D. provided the intellectual basis of the work and designed the experiments. T.T.W., R.M.P., B.H., D.X. and T.V. performed experiments resulting in figures. J.S., R.H. and J.M.H. preformed experiments that contributed intellectually but did not result in figures. R.H. generated reagents. I.M.D. and R.M.P. provided technical expertise for equipment, advanced microscopy and data analysis. T.T.W., R.M.P., C.R. and I.D. wrote and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 2003 kb)
Supplementary Methods
Supplementary Information (PDF 314 kb)
Supplementary Movie 1
Supplementary Information (MOV 7568 kb)
Supplementary Movie 2
Supplementary Information (MOV 491 kb)
Supplementary Movie 3
Supplementary Information (MOV 2370 kb)
Supplementary Movie 4
Supplementary Information (MOV 110 kb)
Supplementary Movie 5
Supplementary Information (MOV 59 kb)
Supplementary Movie 6
Supplementary Information (MOV 213 kb)
Rights and permissions
About this article
Cite this article
Weil, T., Parton, R., Herpers, B. et al. Drosophila patterning is established by differential association of mRNAs with P bodies. Nat Cell Biol 14, 1305–1313 (2012). https://doi.org/10.1038/ncb2627
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb2627
This article is cited by
-
A local translation program regulates centriole amplification in the airway epithelium
Scientific Reports (2023)
-
The PIWI protein Aubergine recruits eIF3 to activate translation in the germ plasm
Cell Research (2020)
-
DEPS-1 is required for piRNA-dependent silencing and PIWI condensate organisation in Caenorhabditis elegans
Nature Communications (2020)
-
mRNA quantification using single-molecule FISH in Drosophila embryos
Nature Protocols (2017)
-
The new (dis)order in RNA regulation
Cell Communication and Signaling (2016)