Abstract
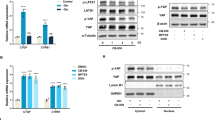
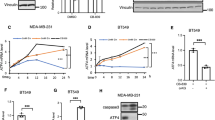
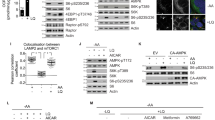
The PI(3)K–PKB–FOXO signalling network provides a major intracellular hub for the regulation of cell proliferation, survival and stress resistance. Here we report an unexpected role for FOXO transcription factors in regulating autophagy by modulating intracellular glutamine levels. To identify transcriptional targets of this network, we performed global transcriptional analyses after conditional activation of the key components PI(3)K, PKB/Akt, FOXO3 and FOXO4. Using this pathway approach, we identified glutamine synthetase as being transcriptionally regulated by PI(3)K–PKB–FOXO signalling. Conditional activation of FOXO also led to an increased level of glutamine production. FOXO activation resulted in mTOR inhibition by preventing the translocation of mTOR to lysosomal membranes in a glutamine-synthetase-dependent manner. This resulted in an increased level of autophagy as measured by LC3 lipidation, p62 degradation and fluorescent imaging of multiple autophagosomal markers. Inhibition of FOXO3-mediated autophagy increased the level of apoptosis, suggesting that the induction of autophagy by FOXO3-mediated glutamine synthetase expression is important for cellular survival. These findings reveal a growth-factor-responsive network that can directly modulate autophagy through the regulation of glutamine metabolism.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Burgering, B. M. & Coffer, P. J. Protein kinase B (c-Akt) in phosphatidylinositol-3-OH kinase signal transduction. Nature 376, 599–602 (1995).
Stephens, L. et al. Protein kinase B kinases that mediate phosphatidylinositol 3,4,5-trisphosphate-dependent activation of protein kinase B. Science 279, 710–714 (1998).
Calnan, D. R. & Brunet, A. The FoxO code. Oncogene 27, 2276–2288 (2008).
Dorman, J. B., Albinder, B., Shroyer, T. & Kenyon, C. The age-1 and daf-2 genes function in a common pathway to control the lifespan of Caenorhabditis elegans. Genetics 141, 1399–1406 (1995).
Kimura, K. D., Tissenbaum, H. A., Liu, Y. & Ruvkun, G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277, 942–946 (1997).
Paradis, S. & Ruvkun, G. Caenorhabditis elegans Akt/PKB transduces insulin receptor-like signals from AGE-1 PI3 kinase to the DAF-16 transcription factor. Genes Dev. 12, 2488–2498 (1998).
Dansen, T. B. & Burgering, B. M. Unravelling the tumor-suppressive functions of FOXO proteins. Trends Cell Biol. 18, 421–429 (2008).
Kops, G. J. et al. Control of cell cycle exit and entry by protein kinase B-regulated forkhead transcription factors. Mol. Cell Biol. 22, 2025–2036 (2002).
Tran, H. et al. DNA repair pathway stimulated by the forkhead transcription factor FOXO3a through the Gadd45 protein. Science 296, 530–534 (2002).
Delpuech, O. et al. Induction of Mxi1-SR alpha by FOXO3a contributes to repression of Myc-dependent gene expression. Mol. Cell Biol. 27, 4917–4930 (2007).
Mei, Y. et al. FOXO3a-dependent regulation of Pink1 (Park6) mediates survival signaling in response to cytokine deprivation. Proc. Natl Acad. Sci. USA 106, 5153–5158 (2009).
van der Vos, K. E. & Coffer, P. J. The extending network of FOXO transcriptional target genes. Antioxid. Redox. Signal. 14, 579–592 (2010).
Eisenberg, D., Gill, H. S., Pfluegl, G. M. & Rotstein, S. H. Structure-function relationships of glutamine synthetases. Biochim. Biophys. Acta 1477, 122–145 (2000).
Berlicki, L. Inhibitors of glutamine synthetase and their potential application in medicine. Mini. Rev. Med. Chem. 8, 869–878 (2008).
DeBerardinis, R.J. & Cheng, T. Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene 29, 313–324 (2010).
Wullschleger, S., Loewith, R. & Hall, M. N. TOR signaling in growth and metabolism. Cell 124, 471–484 (2006).
Sancak, Y. et al. Ragulator–Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303 (2010).
Blommaart, E. F., Luiken, J. J., Blommaart, P. J., van Woerkom, G. M. & Meijer, A. J. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J. Biol. Chem. 270, 2320–2326 (1995).
Mortimore, G. E. & Schworer, C. M. Induction of autophagy by amino-acid deprivation in perfused rat liver. Nature 270, 174–176 (1977).
Kabeya, Y. et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720–5728 (2000).
Rubinsztein, D. C. et al. In search of an ‘autophagomometer’. Autophagy 5, 585–589 (2009).
Proikas-Cezanne, T., Ruckerbauer, S., Stierhof, Y. D., Berg, C. & Nordheim, A. Human WIPI-1 puncta-formation: a novel assay to assess mammalian autophagy. FEBS Lett. 581, 3396–3404 (2007).
Codogno, P., Mehrpour, M. & Proikas-Cezanne, T. Canonical and non-canonical autophagy: variations on a common theme of self-eating? Nat. Rev. Mol. Cell Biol. 13, 7–12 (2011).
Mizushima, N. & Komatsu, M. Autophagy: renovation of cells and tissues. Cell 147, 728–741 (2011).
Lum, J. J. et al. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell 120, 237–248 (2005).
Mizushima, N., Yoshimori, T. & Levine, B. Methods in mammalian autophagy research. Cell 140, 313–326 (2010).
Mammucari, C. et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 6, 458–471 (2007).
Zhao, J. et al. FoxO3 coordinately activates protein degradation by the a utophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 6, 472–483 (2007).
Xu, P., Das, M., Reilly, J. & Davis, R. J. JNK regulates FoxO-dependent autophagy in neurons. Genes Dev. 25, 310–322 (2011).
Zhao, Y. et al. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat. Cell Biol. 12, 665–675 (2010).
Cheong, H., Lindsten, T., Wu, J., Lu, C. & Thompson, C. B. Ammonia-induced autophagy is independent of ULK1/ULK2 kinases. Proc. Natl Acad. Sci. USA 108, 11121–11126 (2011).
Eng, C. H., Yu, K., Lucas, J., White, E. & Abraham, R. T. Ammonia derivedfrom glutaminolysis is a diffusible regulator of autophagy. Sci. Signal. 3, ra31 (2010).
Pattingre, S., Espert, L., Biard-Piechaczyk, M. & Codogno, P. Regulation ofmacroautophagy by mTOR and Beclin 1 complexes. Biochimie 90, 313–323 (2008).
Nicklin, P. et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell 136, 521–534 (2009).
Seglen, P. O. & Gordon, P. B. Amino acid control of autophagic sequestration and protein degradation in isolated rat hepatocytes. J. Cell Biol. 99, 435–444 (1984).
Schworer, C. M. & Mortimore, G. E. Glucagon-induced autophagy and proteolysis in rat liver: mediation by selective deprivation of intracellular amino acids. Proc. Natl Acad. Sci. USA 76, 3169–3173 (1979).
Krause, U., Bertrand, L., Maisin, L., Rosa, M. & Hue, L. Signalling pathways and combinatory effects of insulin and amino acids in isolated rat hepatocytes. Eur. J. Biochem. 269, 3742–3750 (2002).
Gebhardt, R. & Mecke, D. Heterogeneous distribution of glutamine synthetase among rat liver parenchymal cells in situ and in primary culture. EMBO J. 2, 567–570 (1983).
Sakiyama, T., Musch, M. W., Ropeleski, M. J., Tsubouchi, H. & Chang, E. B. Glutamine increases autophagy under Basal and stressed conditions in intestinal epithelial cells. Gastroenterology 136, 924–932 (2009).
Zhao, M. & Klionsky, D. J. AMPK-dependent phosphorylation of ULK1 induces autophagy. Cell Metab. 13, 119–120 (2011).
Tardito, S. et al. The non-proteinogenic amino acids L: -methionine sulfoximine and DL: -phosphinothricin activate mTOR. Amino Acids 42, 2507–2512 (2011).
Paik, J. H. et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell 128, 309–323 (2007).
White, E. & DiPaola, R. S. The double-edged sword of autophagy modulation in cancer. Clin. Cancer Res. 15, 5308–5316 (2009).
Dijkers, P. F., Medema, R. H., Lammers, J. W., Koenderman, L. & Coffer, P. J. Expression of the pro-apoptotic Bcl-2 family member Bim is regulated by the forkhead transcription factor FKHR-L1. Curr. Biol. 10, 1201–1204 (2000).
Dijkers, P. F. et al. Forkhead transcription factor FKHR-L1 modulates cytokine-dependent transcriptional regulation of p27(KIP1). Mol. Cell Biol. 20, 9138–9148 (2000).
Fahrner, J. et al. Identification and functional characterization of regulatory elements of the glutamine synthetase gene from rat liver. Eur. J. Biochem. 213, 1067–1073 (1993).
Gaunitz, F., Weber, S., Scheja, L. & Gebhardt, R. Identification of a cis-acting element and a novel trans-acting factor of the glutamine synthetase gene in liver cells. Biochem. Biophys. Res. Commun. 284, 377–383 (2001).
Pfisterer, S. G., Mauthe, M., Codogno, P. & Proikas-Cezanne, T. Ca2+/calmodulin-dependent kinase (CaMK) signaling via CaMKI and AMP-activated protein kinase contributes to the regulation of WIPI-1 at the onset of autophagy. Mol. Pharmacol. 80, 1066–1075 (2011).
Caldenhoven, E. et al. Activation of the STAT3/acute phase response factor transcription factor by interleukin-5. J. Biol. Chem. 270, 25778–25784 (1995).
Raaben, M. et al. Improved microarray gene expression profiling of virus-infected cells after removal of viral RNA. BMC Genom. 9, 221 (2008).
Deuel, T. F., Louie, M. & Lerner, A. Glutamine synthetase from rat liver. Purification, properties, and preparation of specific antisera. J. Biol. Chem. 253, 6111–6118 (1978).
Guthke, R. et al. Dynamics of amino acid metabolism of primary human liver cells in 3D bioreactors. Bioprocess. Biosyst. Eng. 28, 331–340 (2006).
Brenner, S. The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974).
Lewis, J. A. & Fleming, J. T. Basic culture methods. Methods Cell Biol. 48, 3–29 (1995).
Lin, K., Dorman, J. B., Rodan, A. & Kenyon, C. daf-16: an HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science 278, 1319–1322 (1997).
Acknowledgements
We thank R. Korswagen for helpful discussions. K.E.v.d.V. was supported by a grant from the Dutch Scientific Organisation (NWO-VIDI), R.G. was supported by grants (0313081F and 0315735, Virtual Liver) from the German Federal Ministry of Education and Research (BMBF), T.P-C. was supported by the German Research Foundation (DFG; SFB 773, A03), R.v.B. was supported by a grant from CTMM and L.P.V. was supported by a grant from NIRM.
Author information
Authors and Affiliations
Contributions
K.E.v.d.V. was involved in experimental strategy and design, performed experiments, analysed data and wrote the paper. P.E., S.J.V., R.v.B., M.P., I.J.v.Z., M.M., S.Z., C.P., L.P.V., M.J.A.G.K., A.K.B. and T.B.D. performed experiments. T.P-C., F.C.H., R.G. and B.M.B. were involved in experimental design. P.J.C. was involved in experimental strategy and design and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 539 kb)
Supplementary Table 1
Supplementary Information (XLS 22 kb)
Rights and permissions
About this article
Cite this article
van der Vos, K., Eliasson, P., Proikas-Cezanne, T. et al. Modulation of glutamine metabolism by the PI(3)K–PKB–FOXO network regulates autophagy. Nat Cell Biol 14, 829–837 (2012). https://doi.org/10.1038/ncb2536
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb2536
This article is cited by
-
miRNA-559 and MTDH as possible diagnostic markers of psoriasis: Role of PTEN/AKT/FOXO pathway in disease pathogenesis
Molecular and Cellular Biochemistry (2023)
-
Glutamine synthetase licenses APC/C-mediated mitotic progression to drive cell growth
Nature Metabolism (2022)
-
Co-ingestion of glutamine and leucine synergistically promotes mTORC1 activation
Scientific Reports (2022)
-
FOXO3 Regulates Sevoflurane-Induced Neural Stem Cell Differentiation in Fetal Rats
Cellular and Molecular Neurobiology (2022)
-
Autophagy deficiency in neurodevelopmental disorders
Cell & Bioscience (2021)