Abstract
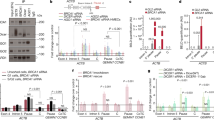
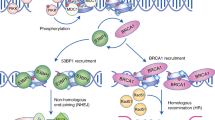
Repair of DNA double-strand breaks is critical to genomic stability and the prevention of developmental disorders and cancer. A central pathway for this repair is homologous recombination (HR). Most knowledge of HR is derived from work in prokaryotic and eukaryotic model organisms. We carried out a genome-wide siRNA-based screen in human cells. Among positive regulators of HR we identified networks of DNA-damage-response and pre-mRNA-processing proteins, and among negative regulators we identified a phosphatase network. Three candidate proteins localized to DNA lesions, including RBMX, a heterogeneous nuclear ribonucleoprotein that has a role in alternative splicing. RBMX accumulated at DNA lesions through multiple domains in a poly(ADP-ribose) polymerase 1-dependent manner and promoted HR by facilitating proper BRCA2 expression. Our screen also revealed that off-target depletion of RAD51 is a common source of RNAi false positives, raising a cautionary note for siRNA screens and RNAi-based studies of HR.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Ciccia, A. & Elledge, S. J. The DNA damage response: making it safe to play with knives. Mol. Cell 40, 179–204 (2010).
Sartori, A. A. et al. Human CtIP promotes DNA end resection. Nature 450, 509–514 (2007).
Bolderson, E. et al. Phosphorylation of Exo1 modulates homologous recombination repair of DNA double-strand breaks. Nucleic Acids Res. 38, 1821–1831 (2010).
West, S. C. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 4, 435–445 (2003).
San Filippo, J., Sung, P. & Klein, H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77, 229–257 (2008).
Heinrich, B. et al. Heterogeneous nuclear ribonucleoprotein G regulates splice site selection by binding to CC(A/C)-rich regions in pre-mRNA. J. Biol. Chem. 284, 14303–14315 (2009).
Pierce, A. J., Johnson, R. D., Thompson, L. H. & Jasin, M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 13, 2633–2638 (1999).
Xia, B. et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol. Cell 22, 719–729 (2006).
Matsuoka, S. et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316, 1160–1166 (2007).
Hurov, K. E., Cotta-Ramusino, C. & Elledge, S. J. A genetic screen identifies the Triple T complex required for DNA damage signaling and ATM and ATR stability. Genes Dev. 24, 1939–1950 (2010).
Paulsen, R. D. et al. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol. Cell 35, 228–239 (2009).
Janknecht, R. Multi-talented DEAD-box proteins and potential tumor promoters: p68 RNA helicase (DDX5) and its paralog, p72 RNA helicase (DDX17). Am. J. Transl. Res. 2, 223–234 (2010).
Lorain, S. et al. Core histones and HIRIP3, a novel histone-binding protein, directly interact with WD repeat protein HIRA. Mol. Cell Biol. 18, 5546–5556 (1998).
Sigoillot, F. D. et al. A bioinformatics method identifies prominent off-targeted transcripts in RNAi screens. Nat. Methods http://dx.doi.org/10.1038/nmeth.1898 (in the press).
Bartel, D. P. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 (2009).
Sigoillot, F. D. & King, R. W. Vigilance and validation: keys to success in RNAi screening. ACS Chem. Biol. 6, 47–60 (2011).
Mazeyrat, S., Saut, N., Mattei, M. G. & Mitchell, M. J. RBMY evolved on the Y chromosome from a ubiquitously transcribed X–Y identical gene. Nat. Genet. 22, 224–226 (1999).
Lingenfelter, P. A. et al. Expression and conservation of processed copies of the RBMX gene. Mamm. Genome 12, 538–545 (2001).
Elliott, D. J. The role of potential splicing factors including RBMY, RBMX, hnRNPG-T and STAR proteins in spermatogenesis. Int. J. Androl. 27, 328–334 (2004).
Shin, K. H., Kang, M. K., Kim, R. H., Christensen, R. & Park, N. H. Heterogeneous nuclear ribonucleoprotein G shows tumor suppressive effect against oral squamous cell carcinoma cells. Clin. Cancer Res. 12, 3222–3228 (2006).
Shin, K. H. et al. p53 promotes the fidelity of DNA end-joining activity by, in part, enhancing the expression of heterogeneous nuclear ribonucleoprotein G. DNA Repair 6, 830–840 (2007).
Chou, D. M. et al. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc. Natl Acad. Sci. USA 107, 18475–18480 (2010).
Polo, S. E., Kaidi, A., Baskcomb, L., Galanty, Y. & Jackson, S. P. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 29, 3130–3139 (2010).
Cotta-Ramusino, C. et al. A DNA damage response screen identifies RHINO, a 9-1-1 and TopBP1 interacting protein required for ATR signaling. Science 332, 1313–1317 (2011).
Smogorzewska, A. et al. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol. Cell 39, 36–47 (2010).
Pierce, A. J., Hu, P., Han, M., Ellis, N. & Jasin, M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 15, 3237–3242 (2001).
Draviam, V. M. et al. A functional genomic screen identifies a role for TAO1 kinase in spindle-checkpoint signalling. Nat. Cell Biol. 9, 556–564 (2007).
Westhorpe, F. G., Diez, M. A., Gurden, M. D., Tighe, A. & Taylor, S. S. Re-evaluating the role of Tao1 in the spindle checkpoint. Chromosoma 119, 371–379 (2010).
Hubner, N. C. et al. Re-examination of siRNA specificity questions role of PICH and Tao1 in the spindle checkpoint and identifies Mad2 as a sensitive target for small RNAs. Chromosoma 119, 149–165 (2010).
Slabicki, M. et al. A genome-scale DNA repair RNAi screen identifies SPG48 as a novel gene associated with hereditary spastic paraplegia. PLoS Biol. 8, e1000408 (2010).
Moumen, A., Masterson, P., O’Connor, M. J. & Jackson, S. P. hnRNP K: an HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell 123, 1065–1078 (2005).
Reinhardt, H. C. et al. DNA damage activates a spatially distinct late cytoplasmic cell-cycle checkpoint network controlled by MK2-mediated RNA stabilization. Mol. Cell 40, 34–49 (2010).
Reinhardt, H. C., Cannell, I. G., Morandell, S. & Yaffe, M. B. Is post-transcriptional stabilization, splicing and translation of selective mRNAs a key to the DNA damage response? Cell Cycle 10, 23–27 (2011).
Bekker-Jensen, S. et al. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J. Cell Biol. 173, 195–206 (2006).
Smogorzewska, A. et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell 129, 289–301 (2007).
Acknowledgements
We thank M. Jasin (Memorial Sloan-Kettering Cancer Center, USA) for DR-U2OS cells, P. Ng and F. Graham (McMaster University, Canada) for the AdNGUS24i, P. D. Adams (Institute of Cancer Sciences, CR-UK Beatson Labs, UK) for antibodies, S. J. Boulton (DNA Damage Response Laboratory, London Research Institute, UK) for the PARP inhibitor and C. Cotta-Ramusino and members of the Elledge laboratory for advice and discussion. We thank the Institute of Chemistry and Cell Biology (ICCB)-Longwood screening facility, including C. Shamu, S. Rudnicki, S. M. Johnston and T. Xie. This work was supported by a grant from the National Institutes of Health to S.J.E. A.S. was supported by T32CA09216 to the Pathology Department at the Massachusetts General Hospital and by Burroughs Wellcome Fund Career Award for Medical Scientists and is a Rita Allen Foundation and an Irma T. Hirschl scholar. S.J.E. is an investigator with the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
B.A., A.S. and S.J.E. conceived experimental design and conducted data analysis. F.D.S. and R.W.K. carried out GESS analysis. The manuscript was prepared by B.A. and S.J.E. and edited by A.S., F.D.S. and R.W.K.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 4811 kb)
Supplementary Table 1
Supplementary Information (XLSX 2365 kb)
Supplementary Table 2
Supplementary Information (XLSX 373 kb)
Supplementary Table 3
Supplementary Information (XLSX 167 kb)
Supplementary Table 4
Supplementary Information (XLSX 220 kb)
Supplementary Table 5
Supplementary Information (XLSX 37 kb)
Supplementary Table 6
Supplementary Information (XLSX 43 kb)
Supplementary Table 7
Supplementary Information (XLSX 40 kb)
Supplementary Table 8
Supplementary Information (XLSX 59 kb)
Supplementary Table 9
Supplementary Information (XLSX 52 kb)
Supplementary Table 10
Supplementary Information (XLSX 56 kb)
Rights and permissions
About this article
Cite this article
Adamson, B., Smogorzewska, A., Sigoillot, F. et al. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat Cell Biol 14, 318–328 (2012). https://doi.org/10.1038/ncb2426
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb2426
This article is cited by
-
Integrative modeling of lncRNA-chromatin interaction maps reveals diverse mechanisms of nuclear retention
BMC Genomics (2023)
-
Racial differences in RAD51 expression are regulated by miRNA-214-5P and its inhibition synergizes with olaparib in triple-negative breast cancer
Breast Cancer Research (2023)
-
PDHB-AS suppresses cervical cancer progression and cisplatin resistance via inhibition on Wnt/β-catenin pathway
Cell Death & Disease (2023)
-
A dual role of RBM42 in modulating splicing and translation of CDKN1A/p21 during DNA damage response
Nature Communications (2023)
-
Recognition of the Subtypes Classification and Diagnostic Signature Based on RNA N6-Methyladenosine Regulators in Recurrent Spontaneous Abortion
Reproductive Sciences (2023)