Abstract
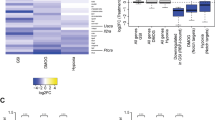
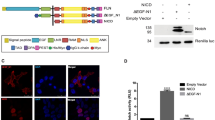
Cellular decisions of self-renewal or differentiation arise from integration and reciprocal titration of numerous regulatory networks. Notch and Wnt/β-catenin signalling often intersect in stem and progenitor cells and regulate each other transcriptionally. The biological outcome of signalling through each pathway often depends on the context and timing as cells progress through stages of differentiation. Here, we show that membrane-bound Notch physically associates with unphosphorylated (active) β-catenin in stem and colon cancer cells and negatively regulates post-translational accumulation of active β-catenin protein. Notch-dependent regulation of β-catenin protein did not require ligand-dependent membrane cleavage of Notch or the glycogen synthase kinase- 3β-dependent activity of the β-catenin destruction complex. It did, however, require the endocytic adaptor protein Numb and lysosomal activity. This study reveals a previously unrecognized function of Notch in negatively titrating active β-catenin protein levels in stem and progenitor cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Logan, C. Y. & Nusse, R. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 20, 781–810 (2004).
Radtke, F. & Clevers, H. Self-renewal and cancer of the gut: two sides of a coin. Science 307, 1904–1909 (2005).
Hayward, P., Kalmar, T. & Arias, A. M. Wnt/Notch signalling and information processing during development. Development 135, 411–424 (2008).
Artavanis-Tsakonas, S., Rand, M. D. & Lake, R. J. Notch signaling: cell fate control and signal integration in development. Science 284, 770–776 (1999).
Kwon, C. et al. A regulatory pathway involving Notch1/ β-catenin/Isl1 determines cardiac progenitor cell fate. Nat. Cell Biol. 11, 951–957 (2009).
van Noort, M., Weerkamp, F., Clevers, H. C. & Staal, F. J. Wnt signaling and phosphorylation status of β-catenin: importance of the correct antibody tools. Blood 110, 2778–2779 (2007).
Nemir, M., Croquelois, A., Pedrazzini, T. & Radtke, F. Induction of cardiogenesis in embryonic stem cells via downregulation of Notch1 signaling. Circ. Res. 98, 1471–1478 (2006).
Yang, X. et al. Notch activation induces apoptosis in neural progenitor cells through a p53-dependent pathway. Dev. Biol. 269, 81–94 (2004).
Tanigaki, K. et al. Notch-RBP-J signaling is involved in cell fate determination of marginal zone B cells. Nat. Immunol. 3, 443–450 (2002).
Srinivas, S. et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1, 4 (2001).
Cai, C. L. et al. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev. Cell 5, 877–889 (2003).
Kwon, C. et al. Canonical Wnt signaling is a positive regulator of mammalian cardiac progenitors. Proc. Natl Acad. Sci. USA 104, 10894–10899 (2007).
Qyang, Y. et al. The renewal and differentiation of Isl1+ cardiovascularprogenitors are controlled by a Wnt/β-catenin pathway. Cell Stem Cell 1, 165–179 (2007).
Martinez Arias, A., Zecchini, V. & Brennan, K. CSL-independent Notch signalling: a checkpoint in cell fate decisions during development? Curr. Opin. Genet. Dev. 12, 524–533 (2002).
Yan, D. et al. Elevated expression of axin2 and hnkd mRNA provides evidence that Wnt/ β -catenin signaling is activated in human colon tumors. Proc. Natl Acad. Sci. USA 98, 14973–14978 (2001).
Tetsu, O. & McCormick, F. β-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398, 422–426 (1999).
Meijer, L. et al. GSK-3-selective inhibitors derived from Tyrian purple indirubins. Chem. Biol. 10, 1255–1266 (2003).
McElhinny, A. S., Li, J. L. & Wu, L. Mastermind-like transcriptional co-activators: emerging roles in regulating cross talk among multiple signaling pathways. Oncogene 27, 5138–5147 (2008).
Harada, N. et al. Intestinal polyposis in mice with a dominant stable mutation of the β-catenin gene. EMBO J. 18, 5931–5942 (1999).
Murtaugh, L. C., Stanger, B. Z., Kwan, K. M. & Melton, D. A. Notch signaling controls multiple steps of pancreatic differentiation. Proc. Natl Acad. Sci. USA 100, 14920–14925 (2003).
Korinek, V. et al. Constitutive transcriptional activation by a β-catenin–Tcf complex in APC-/- colon carcinoma. Science 275, 1784–1787 (1997).
Yamamoto, N. et al. Role of Deltex-1 as a transcriptional regulator downstream of the Notch receptor. J. Biol. Chem. 276, 45031–45040 (2001).
Tamura, K. et al. Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-J κ/Su(H). Curr. Biol. 5, 1416–1423 (1995).
Schroeter, E. H., Kisslinger, J. A. & Kopan, R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature 393, 382–386 (1998).
Lindsley, R. C., Gill, J. G., Kyba, M., Murphy, T. L. & Murphy, K. M. Canonical Wnt signaling is required for development of embryonic stem cell-derived mesoderm. Development 133, 3787–3796 (2006).
Fehling, H. J. et al. Tracking mesoderm induction and its specification to the hemangioblast during embryonic stem cell differentiation. Development 130, 4217–4227 (2003).
De Strooper, B. et al. A presenilin-1-dependent γ-secretase-like protease mediates release of Notch intracellular domain. Nature 398, 518–522 (1999).
Sastre, M. et al. Presenilin-dependent γ-secretase processing of β-amyloid precursor protein at a site corresponding to the S3 cleavage of Notch. EMBO Rep. 2, 835–841 (2001).
Ge, C. & Stanley, P. Effects of varying Notch1 signal strength on embryogenesis and vasculogenesis in compound mutant heterozygotes. BMC Dev. Biol. 10, 36 (2010).
Kanwar, R. & Fortini, M. E. Notch signaling: a different sort makes the cut. Curr. Biol. 14, R1043–R1045 (2004).
Guo, M., Jan, L. Y. & Jan, Y. N. Control of daughter cell fates during asymmetric division: interaction of Numb and Notch. Neuron 17, 27–41 (1996).
Zhong, W., Jiang, M. M., Weinmaster, G., Jan, L. Y. & Jan, Y. N. Differential expression of mammalian Numb, Numblike and Notch1 suggests distinct roles during mouse cortical neurogenesis. Development 124, 1887–1897 (1997).
McGill, M. A., Dho, S. E., Weinmaster, G. & McGlade, C. J. Numb regulates post-endocytic trafficking and degradation of Notch1. J. Biol. Chem. 284, 26427–26438 (2009).
Tapper, H. & Sundler, R. Bafilomycin A1 inhibits lysosomal, phagosomal, and plasma membrane PH(+)-ATPase and induces lysosomal enzyme secretion in macrophages. J. Cell Physiol. 163, 137–144 (1995).
Chan, A. T. et al. Long-term use of aspirin and nonsteroidal anti-inflammatory drugs and risk of colorectal cancer. JAMA 294, 914–923 (2005).
Rostom, A. et al. Nonsteroidal anti-inflammatory drugs and cyclooxygenase-2 inhibitors for primary prevention of colorectal cancer: a systematic review prepared for the U.S. Preventive Services Task Force. Ann. Intern. Med. 146, 376–389 (2007).
Eriksen, J. L. et al. NSAIDs and enantiomers of flurbiprofen target γ-secretase and lower A β 42 in vivo. J. Clin. Invest. 112, 440–449 (2003).
Koch, U. & Radtke, F. Notch and cancer: a double-edged sword. Cell Mol. Life Sci. 64, 2746–2762 (2007).
van Es, J. H. et al. Notch/ γ-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature 435, 959–963 (2005).
Sanders, P. G. et al. Ligand-independent traffic of Notch buffers activated Armadillo in Drosophila. PLoS Biol. 7, e1000169 (2009).
Acknowledgements
We thank R. Kopan (Washington University), M. Nakafuku (Cincinnati Children’s Hospital), T. Honjo (Kyoto University) and P. Stanley (Albert Einstein College of Medicine) for providing tethered Notch constructs, Notch deletion constructs, RBP-Jflox mice and Notch1lbd/lbd ES cells, respectively. The authors thank G. Howard and S. Ordway for editorial assistance, Srivastava and Kwon laboratory members for discussions, B. Taylor for assistance with manuscript and figure preparation and B. Bruneau for critical reading of the manuscript. We also thank J. Fish and C. Miller in the Gladstone Histology core. C.K. was supported by grants from the American Heart Association Beginning Grant-in-Aid and National Heart, Lung, and Blood Institute/National Institutes of Health (NHLBI/NIH; 1K99HL092234, 4R00HL09223); I.N.K. was supported by a March of Dimes Basil O’Connor Award; D.S. was supported by grants from NHLBI/NIH (P01 HL089707, U01 HL100406), the California Institute for Regenerative Medicine and the Younger Family Foundation. This work was supported by NIH/National Center for Research Resources grant C06 RR018928 to the Gladstone Institute.
Author information
Authors and Affiliations
Contributions
C.K. designed, carried out and supervised in vivo and in vitro work and wrote the manuscript. P.C. designed and carried out in vivo and in vitro work and wrote the manuscript. I.N.K. carried out Notch Co-IP and western analyses. P.A. cultured embryonic stem cells and carried out luciferase assays. L.S. carried out immunocytochemistry and confocal microscopy. V.N. isolated mesenchymal stem cells and carried out western analyses. D.S. designed and supervised this work and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
D.S. is a scientific co-founder of iPierian Inc. and is a member of the Scientific Advisory Board of iPierian Inc. and RegeneRx Pharmaceuticals.
Supplementary information
Supplementary Information
Supplementary Information (PDF 707 kb)
Rights and permissions
About this article
Cite this article
Kwon, C., Cheng, P., King, I. et al. Notch post-translationally regulates β-catenin protein in stem and progenitor cells. Nat Cell Biol 13, 1244–1251 (2011). https://doi.org/10.1038/ncb2313
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb2313
This article is cited by
-
Oral squamous cell carcinomas: state of the field and emerging directions
International Journal of Oral Science (2023)
-
WNT ligands in non-small cell lung cancer: from pathogenesis to clinical practice
Discover Oncology (2023)
-
Notch ankyrin domain: evolutionary rise of a thermodynamic sensor
Cell Communication and Signaling (2022)
-
Wnt signaling in colorectal cancer: pathogenic role and therapeutic target
Molecular Cancer (2022)
-
Wnt/β-catenin signalling: function, biological mechanisms, and therapeutic opportunities
Signal Transduction and Targeted Therapy (2022)