Abstract
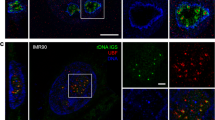
The cell nucleus is a highly compartmentalized organelle harbouring a variety of dynamic membraneless nuclear bodies1,2,3,4. How these subnuclear domains are established and maintained is not well understood5,6,7,8. Here, we investigate the molecular mechanism of how one nuclear body, the paraspeckle, is assembled and organized. Paraspeckles are discrete ribonucleoprotein bodies found in mammalian cells and implicated in nuclear retention of hyperedited mRNAs9,10,11. We developed a live-cell imaging system that allows for the inducible transcription of Men ɛ/β (also known as Neat1; ref. 12) noncoding RNAs (ncRNAs) and the direct visualization of the recruitment of paraspeckle proteins. Using this system, we demonstrate that Men ɛ/β ncRNAs are essential to initiate the de novo assembly of paraspeckles. These newly formed structures effectively harbour nuclear-retained mRNAs confirming that they are bona fide functional paraspeckles. By three independent approaches, we show that it is the act of Men ɛ/β transcription, but not ncRNAs alone, that regulates paraspeckle maintenance. Finally, fluorescence recovery after photobleaching (FRAP) analyses supported a critical structural role for Men ɛ/β ncRNAs in paraspeckle organization. This study establishes a model in which Men ɛ/β ncRNAs serve as a platform to recruit proteins to assemble paraspeckles.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Handwerger, K. E. & Gall, J. G. Subnuclear organelles: new insights into form and function. Trends Cell Biol. 16, 19–26 (2006).
Lamond, A. I. & Spector, D. L. Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 4, 605–612 (2003).
Spector, D. L. The dynamics of chromosome organization and gene regulation. Annu. Rev. Biochem. 72, 573–608 (2003).
Zhao, R., Bodnar, M. S. & Spector, D. L. Nuclear neighborhoods and gene expression. Curr. Opin. Genet. Dev. 19, 172–179 (2009).
Kumaran, R. I., Thakar, R. & Spector, D. L. Chromatin dynamics and gene positioning. Cell 132, 929–934 (2008).
Matera, A. G., Izaguire-Sierra, M., Praveen, K. & Rajendra, T. K. Nuclear bodies: random aggregates of sticky proteins or crucibles of macromolecular assembly? Dev. Cell 17, 639–647 (2009).
Misteli, T. The concept of self-organization in cellular architecture. J. Cell Biol. 155, 181–185 (2001).
Misteli, T. Beyond the sequence: cellular organization of genome function. Cell 128, 787–800 (2007).
Chen, L. L., DeCerbo, J. N. & Carmichael, G. G. Alu element-mediated gene silencing. EMBO J. 27, 1694–1705 (2008).
Prasanth, K. V. et al. Regulating gene expression through RNA nuclear retention. Cell 123, 249–263 (2005).
Zhang, Z. & Carmichael, G. G. The fate of dsRNA in the nucleus: a p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell 106, 465–475 (2001).
Hutchinson, J. N. et al. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 8, 39 (2007).
Kaiser, T. E., Intine, R. V. & Dundr, M. De novo formation of a subnuclear body. Science 322, 1713–1717 (2008).
Bond, C. S. & Fox, A. H. Paraspeckles: nuclear bodies built on long noncoding RNA. J. Cell Biol. 186, 637–644 (2009).
Fox, A. H., Bond, C. S. & Lamond, A. I. P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol. Biol. Cell 16, 5304–5315 (2005).
Chen, L. L. & Carmichael, G. G. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol. Cell 35, 467–478 (2009).
Clemson, C. M. et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 33, 717–726 (2009).
Sasaki, Y. T., Ideue, T., Sano, M., Mituyama, T. & Hirose, T. MEN ɛ/β noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc. Natl Acad. Sci. USA 106, 2525–2530 (2009).
Sunwoo, H. et al. MEN ɛ/β nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 19, 347–359 (2009).
Kumaran, R. I. & Spector, D. L. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J. Cell Biol. 180, 51–65 (2008).
Janicki, S. M. et al. From silencing to gene expression: real-time analysis in single cells. Cell 116, 683–698 (2004).
Tsukamoto, T. et al. Visualization of gene activity in living cells. Nat. Cell Biol. 2, 871–878 (2000).
Bertrand, E. et al. Localization of ASH1 mRNA particles in living yeast. Mol. Cell 2, 437–445 (1998).
Fusco, D. et al. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr. Biol. 13, 161–167 (2003).
Sasaki, Y. T. & Hirose, T. How to build a paraspeckle. Genome Biol. 10, 227 (2009).
Dousset, T. et al. Initiation of nucleolar assembly is independent of RNA polymerase I transcription. Mol. Biol. Cell 11, 2705–2717 (2000).
Cardinale, S. et al. Subnuclear localization and dynamics of the pre-mRNA 3′ end processing factor mammalian cleavage factor I 68-kDa subunit. Mol. Biol. Cell 18, 1282–1292 (2007).
Zhang, W. W., Zhang, L. X., Busch, R. K., Farres, J. & Busch, H. Purification and characterization of a DNA-binding heterodimer of 52 and 100 kDa from HeLa cells. Biochem. J. 290, 267–272 (1993).
Shav-Tal, Y. et al. Dynamics of single mRNPs in nuclei of living cells. Science 304, 1797–1800 (2004).
Acknowledgements
We thank J. Caceres, G. G. Carmichael, L.-L. Chen, Y. Kurihara, and A. I. Lamond for reagents, S. Hearn and Z. Lazar for assistance in microscopy, C. Berasain, M. Bodnar, M. Eckersley-Maslin, M. Huebner, I. R. Kumaran, J. Li, E. Reis, J. E. Wilusz, and R. Zhao of the Spector laboratory for discussions and comments. Y.S.M. is supported by a National Cancer Center Postdoctoral Fellowship. B.Z. is supported by a Department of Defense Prostate Cancer Research Program Postdoctoral Fellowship. This work was supported by grants to D.L.S. from NIH (NIGMS 42694 and 5PO1CA013106-38).
Author information
Authors and Affiliations
Contributions
Y.S.M., H.S., and D.L.S. designed the experiments. Y.S.M., H.S., and B.Z. performed experiments and analysed data. Y.S.M. and D.L.S. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 875 kb)
Supplementary Movie 1
Supplementary Information (MPG 1376 kb)
Supplementary Movie 2
Supplementary Information (MPG 2825 kb)
Supplementary Movie 3
Supplementary Information (MPG 427 kb)
Supplementary Movie 4
Supplementary Information (MPG 258 kb)
Supplementary Movie 5
Supplementary Information (MPG 309 kb)
Supplementary Movie 6
Supplementary Information (MPG 112 kb)
Supplementary Movie 7
Supplementary Information (MPG 347 kb)
Supplementary Movie 8
Supplementary Information (MPG 3566 kb)
Supplementary Movie 9
Supplementary Information (MPG 1068 kb)
Supplementary Movie 10
Supplementary Information (MPG 415 kb)
Supplementary Movie 11
Supplementary Information (MPG 299 kb)
Rights and permissions
About this article
Cite this article
Mao, Y., Sunwoo, H., Zhang, B. et al. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol 13, 95–101 (2011). https://doi.org/10.1038/ncb2140
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb2140
This article is cited by
-
Investigating phase separation properties of chromatin-associated proteins using gradient elution of 1,6-hexanediol
BMC Genomics (2023)
-
Shell protein composition specified by the lncRNA NEAT1 domains dictates the formation of paraspeckles as distinct membraneless organelles
Nature Cell Biology (2023)
-
A model for organization and regulation of nuclear condensates by gene activity
Nature Communications (2023)
-
VGLL3 is a mechanosensitive protein that promotes cardiac fibrosis through liquid–liquid phase separation
Nature Communications (2023)
-
From genotype to phenotype: genetics of mammalian long non-coding RNAs in vivo
Nature Reviews Genetics (2022)