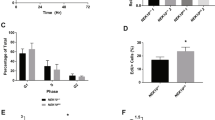
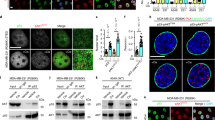
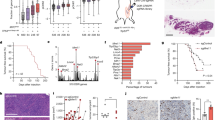
Abstract
Cells can undergo either cell-cycle arrest or apoptosis after genotoxic stress, based on p53 activity1,2,3,4,5,6. Here we show that cellular fate commitment depends on Axin forming distinct complexes with Pirh2, Tip60, HIPK2 and p53. In cells treated with sublethal doses of ultra-violet (UV) radiation or doxorubicin (Dox), Pirh2 abrogates Axin-induced p53 phosphorylation at Ser 46 catalysed by HIPK2, by competing with HIPK2 for binding to Axin. However, on lethal treatment, Tip60 interacts with Axin and abrogates Pirh2–Axin binding, forming an Axin–Tip60–HIPK2–p53 complex that allows maximal p53 activation to trigger apoptosis. We also provide evidence that the ATM/ATR pathway mediates the Axin–Tip60 complex assembly. An axin mutation promotes carcinogenesis in AxinFu/+ (Axin-Fused) mice, consistent with a dominant-negative role for AxinFu in p53 activation. Thus, Axin is a critical determinant in p53-dependent tumour suppression in which Pirh2 and Tip60 have different roles in triggering cell-cycle arrest or apoptosis depending on the severity of genotoxic stress.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
28 August 2009
In the version of this article initially published, the word Tip60 was incorrectly used instead of Pirh2 in the legend for Fig. 2b. This error has been corrected in the HTML and PDF versions of the article.
References
Riley, T., Sontag, E., Chen, P. & Levine, A. Transcriptional control of human p53-regulated genes. Nature Rev. Mol. Cell Biol. 9, 402–412 (2008).
Masha, V., Poyurovsky & Prives, C. Unleashing the power of p53: lessons from mice and men. Genes Dev. 20, 125–131 (2006).
Vousden, K. H. & Lu, X. Live or let die: the cell's response to p53. Nature. Rev. Cancer 8, 594–604 (2002).
Dey, A., Verma, C. S. & Lane, D. P. Updates on p53: modulation of p53 degradation as a therapeutic approach. Br. J. Cancer 98, 4–8 (2008).
Oren, M. Decision making by p53: life, death and cancer. Cell Death Differ. 4, 431–442 (2003).
Harper, J. W. & Elledge, S. J. The DNA damage response: ten years after. Mol. Cell 28, 739–745 (2007).
D'Orazi, G. et al. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser-46 and mediates apoptosis. Nature Cell Biol. 4, 11–19 (2002).
Hofmann, T. G. et al. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nature Cell Biol. 4, 1–10 (2002).
Rinaldo, C. et al. MDM2-regulated degradation of HIPK2 prevents p53 Ser46 phosphorylation and DNA damage-induced apoptosis. Mol. Cell 9, 739–750 (2007).
Gresko, E. et al. Autoregulatory control of the p53 response by caspase-mediated processing of HIPK2. EMBO J. 25, 1883–1894 (2006).
Oda, K. et al. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell 102, 849–862 (2000).
Polakis, P. The many ways of Wnt in cancer. Curr. Opin. Genet. Dev. 17, 45–51 (2007).
Zeng, L. et al. The mouse Fused locus encodes Axin, an inhibitor of the Wnt signaling pathway that regulates embryonic axis formation. Cell 90, 181–192 (1997).
Inoki, K. et al. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell 126, 955–68 (2006).
Rui, Y. et al. A β-catenin-independent dorsalization pathway activated by Axin/JNK signaling and antagonized by Aida. Dev. Cell 13, 268–82 (2007).
Wan, M. et al. Parathyroid hormone signaling through low-density lipoprotein-related protein 6. Genes Dev. 21, 2968–2979 (2008).
Rui, Y. et al. Axin stimulates p53 functions by activation of HIPK2 kinase through multimeric complex formation. EMBO J. 23, 4583–4594 (2004).
Li, Q. et al. Daxx cooperates with the Axin/HIPK2/p53 complex to induce cell death. Cancer Res. 67, 66–74 (2007).
Lu, Z. et al. Protein encoded by the Axin (Fu) allele effectively down-regulates Wnt signaling but exerts a dominant negative effect on c-Jun N-terminal kinase signaling. J. Biol. Chem. 283, 13132–13139 (2008).
Sun, P. et al. PRAK is essential for ras-induced senescence and tumor suppression. Cell 128, 295–308 (2007).
Leng, R. P. et al. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell 112, 779–791 (2003).
Beitel, L. K. et al. Cloning and characterization of an androgen receptor N-terminal-interacting protein with ubiquitin-protein ligase activity. J. Mol. Endocrinol. 29, 41–60 (2002).
Hattori, T. et al. Pirh2 promotes ubiquitin-dependent degradation of the cyclin-dependent kinase inhibitor p27Kip1. Cancer Res. 67, 10789–10795 (2007).
Legube, G. et al. Role of the histone acetyl transferase Tip60 in the p53 pathway. J Biol Chem. 279, 44825–44833 (2004).
Tang, Y., Luo, J., Zhang, W. & Gu, W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol. Cell 24, 827–839 (2006).
Sykes, S. M. et al. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol. Cell 24, 841–851 (2006).
Winter, M., Sombroek, D., Dauth, I., Moehlenbrink, J., Scheuermann, K., Crone, J. & Hofmann, T. G. Control of HIPK2 stability by ubiquitin ligase Siah-1 and checkpoint kinases ATM and ATR. Nature Cell Biol. 10, 812–824 (2008).
Brummelkamp, T. R., Bernards, R. & Agami, R. A system for stable expression of short interfering RNAs in mammalian cells. Science 296, 550–553 (2002).
Harada, J. et al. Requirement of the co-repressor homeodomain-interacting protein kinase 2 for ski-mediated inhibition of bone morphogenetic protein-induced transcriptional activation. J. Biol. Chem. 278, 38998–39005 (2003).
Acknowledgements
This work was supported by grants from the 973 Program and 863 Program (2007CB914602, 2006CB503900 and 2006AA02A303), the National Natural Science Foundation of China (30730025, 30871280 and 30770454), the Ministry of Education of China (705030 and B06016), the National Basic Research Program of the Ministry of Science and Technology and the National Science Foundation of Fujian Province (2009J06021). We thank H. You for commenting on the manuscript.
Author information
Authors and Affiliations
Contributions
Q.L., S.L. and S.C.L. conceived and designed the study. Q.L., S.L., X.W., G.L., Z.L., H.G. and R.K. performed the research; Y.W., Z.Y. and J.H. helped with data analysis and Q.L., S.L. and S.C.L. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 1870 kb)
Rights and permissions
About this article
Cite this article
Li, Q., Lin, S., Wang, X. et al. Axin determines cell fate by controlling the p53 activation threshold after DNA damage. Nat Cell Biol 11, 1128–1134 (2009). https://doi.org/10.1038/ncb1927
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb1927
This article is cited by
-
The scaffold protein AXIN1: gene ontology, signal network, and physiological function
Cell Communication and Signaling (2024)
-
Low glucose metabolite 3-phosphoglycerate switches PHGDH from serine synthesis to p53 activation to control cell fate
Cell Research (2023)
-
p53 regulation by ubiquitin and ubiquitin-like modifications
Genome Instability & Disease (2022)
-
TIP60 recruits SUV39H1 to chromatin to maintain heterochromatin genome stability and resist hydrogen peroxide-induced cytotoxicity
Genome Instability & Disease (2020)
-
E3 ubiquitin ligases in cancer and implications for therapies
Cancer and Metastasis Reviews (2017)