Abstract
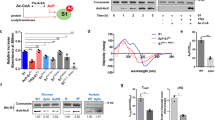
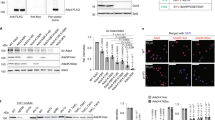
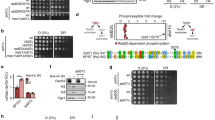
The SNF2h (sucrose non-fermenting protein 2 homologue)-containing chromatin-remodelling complex NoRC silences a fraction of ribosomal RNA genes (rDNA) by establishing a heterochromatic structure at the rDNA promoter1,2,3. Here we show that the acetyltransferase MOF (males absent on the first) acetylates TIP5, the largest subunit of NoRC, at a single lysine residue, K633, adjacent to the TIP5 RNA-binding domain, and that the NAD+-dependent deacetylase SIRT1 (sirtuin-1) removes the acetyl group from K633. Acetylation regulates the interaction of NoRC with promoter-associated RNA (pRNA), which in turn affects heterochromatin formation, nucleosome positioning and rDNA silencing. Significantly, NoRC acetylation is responsive to the intracellular energy status and fluctuates during S phase. Activation of SIRT1 on glucose deprivation leads to deacetylation of K633, enhanced pRNA binding and an increase in heterochromatic histone marks. These results suggest a mechanism that links the epigenetic state of rDNA to cell metabolism and reveal another layer of epigenetic control that involves post-translational modification of a chromatin remodelling complex.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Strohner, R. et al. NoRC - a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J. 20, 4892–4900 (2001).
Zhou, Y., Santoro, R. & Grummt, I. The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription. EMBO J. 21, 4632–4640 (2002).
Santoro, R., Li, J. & Grummt, I. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nature Genet. 32, 393–396 (2002).
Corona, D. F., Clapier, C. R., Becker, P. B. & Tamkun, J. W. Modulation of ISWI function by site-specific histone acetylation. EMBO Rep. 3, 242–247 (2002).
Kurdistani, S. K., Tavazoie, S. & Grunstein, M. Mapping global histone acetylation patterns to gene expression. Cell 117, 721–733 (2004).
Smith, E. R. et al. The Drosophila MSL complex acetylates histone H4 at lysine 16, a chromatin modification linked to dosage compensation. Mol. Cell. Biol. 20, 312–318 (2000).
Akhtar, A. & Becker, P. B. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol. Cell 5, 367–375 (2000).
Lee, K. K. & Workman, J. L. Histone acetyltransferase complexes: one size doesn't fit all. Nature Rev. Mol. Cell Biol. 8, 284–295 (2007).
Dou, Y. et al. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell 121, 873–885 (2005).
Hilfiker, A., Hilfiker-Kleiner, D., Pannuti, A. & Lucchesi, J. C. MOF, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. EMBO J. 16, 2054–2060 (1997).
Buscaino, A. et al. MOF-regulated acetylation of MSL-3 in the Drosophila dosage compensation complex. Mol. Cell 11, 1265–1277 (2003). .
Zhou, Y. & Grummt, I. The PHD finger/bromodomain of NoRC interacts with acetylated histone H4K16 and is sufficient for rDNA silencing. Curr. Biol. 15, 1434–1438 (2005).
Li, J., Längst, G. & Grummt, I. NoRC-dependent nucleosome positioning silences rRNA genes. EMBO J. 25, 5735–5741 (2006).
Taipale, M. et al. MOF histone acetyltransferase is required for histone H4 lysine 16 acetylation in mammalian cells. Mol. Cell. Biol. 25, 6798–6810 (2005).
Gupta, A. et al. Involvement of human MOF in ATM function. Mol. Cell. Biol. 25, 5292–5305 (2005).
Smith, E. R. et al. A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Mol. Cell. Biol. 25, 9175–9188 (2005).
Mendjan, S. et al. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol. Cell 21, 811–823 (2006).
Mayer, C., Schmitz, K. M., Li, J., Grummt, I. & Santoro, R. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol. Cell 22, 351–361 (2006).
Mayer, C., Neubert, M. & Grummt, I. The structure of NoRC-associated RNA is critical for targeting the chromatin remodeling complex NoRC to the nucleolus. EMBO Rep. 9, 774–778 (2008).
Rodgers, J. T. et al. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature 434, 113–118 (2005).
Fulco, M. et al. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev. Cell 14, 661–673 (2008).
Li, J., Santoro, R., Koberna, K. & Grummt, I. The chromatin remodeling complex NoRC controls replication timing of rRNA genes. EMBO J. 24, 120–127 (2004).
Santoro, R. & Grummt, I. Molecular mechanisms mediating methylation-dependent silencing of ribosomal gene transcription. Mol. Cell 8, 719–725 (2001).
Murayama, A. et al. Epigenetic control of rDNA loci in response to intracellular energy status. Cell 133, 627–639 (2008).
Grummt, I. & Ladurner, A. G. A metabolic throttle regulates the epigenetic state of rDNA. Cell 133, 577–580 (2008).
Frye, R. A. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem. Biophys. Res. Commun. 260, 273–279 (1999).
Ramirez-Carrozzi, V. R. et al. Selective and antagonistic functions of SWI/SNF and Mi-2β nucleosome remodeling complexes during an inflammatory response. Genes Dev. 20, 282–296 (2006).
Schmitz, K. M. et al. TAF12 recruits Gadd45a and the nucleotide excision repair complex to the promoter of rRNA genes leading to active DNA demethylation. Mol Cell 33, 344–353 (2009).
Acknowledgements
We thank S. Smale for providing the vector pQXCIP, L. Guarente for expression vectors encoding SIRT1H355Y, G. Xouri and M. Gentzel for their contribution at the early stages of this work and M. Wilm for his support in the mass spectroscopic analysis. This study was funded by the Deutsche Forschungsgemeinschaft (SFB/Transregio 5, SP 'Epigenetics'), the EU-Network 'Epigenome' and the Fonds der Chemischen Industrie.
Author information
Authors and Affiliations
Contributions
Y.G., K.S., C.M. and I.G. conceived the experiments and wrote the manuscript. Y.G., K.S., C.M. and X.Y. performed and analysed the experiments and generated the figures. A.A. contributed the reagents and materials.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 862 kb)
Rights and permissions
About this article
Cite this article
Zhou, Y., Schmitz, KM., Mayer, C. et al. Reversible acetylation of the chromatin remodelling complex NoRC is required for non-coding RNA-dependent silencing. Nat Cell Biol 11, 1010–1016 (2009). https://doi.org/10.1038/ncb1914
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb1914
This article is cited by
-
The AT-hook is an evolutionarily conserved auto-regulatory domain of SWI/SNF required for cell lineage priming
Nature Communications (2023)
-
Aberrant (pro)renin receptor expression induces genomic instability in pancreatic ductal adenocarcinoma through upregulation of SMARCA5/SNF2H
Communications Biology (2020)
-
Nucleolus and chromatin
Histochemistry and Cell Biology (2018)
-
Functions of bromodomain-containing proteins and their roles in homeostasis and cancer
Nature Reviews Molecular Cell Biology (2017)
-
The PML nuclear bodies-associated protein TTRAP regulates ribosome biogenesis in nucleolar cavities upon proteasome inhibition
Cell Death & Differentiation (2012)