Abstract
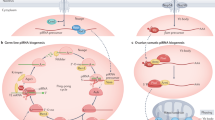
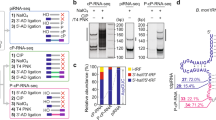
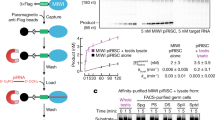
Piwi family proteins are essential for germline development and bind piwi-interacting RNAs (piRNAs)1,2,3. The grandchildless gene aub of Drosophila melanogaster encodes the piRNA-binding protein Aubergine (Aub), which is essential for formation of primordial germ cells (PGCs)4. Here we report that Piwi family proteins of mouse, Xenopus laevis and Drosophila contain symmetrical dimethylarginines (sDMAs). We found that Piwi proteins are expressed in Xenopus oocytes and we identified numerous Xenopus piRNAs. We report that the Drosophila homologue of protein methyltransferase 5 (dPRMT5, csul/dart5), which is also the product of a grandchildless gene5,6, is required for arginine methylation of Drosophila Piwi, Ago3 and Aub proteins in vivo. Loss of dPRMT5 activity led to a reduction in the levels of piRNAs, Ago3 and Aub proteins, and accumulation of retrotransposons in the Drosophila ovary. Our studies explain the relationship between aub and dPRMT5 (csul/dart5) genes by demonstrating that dPRMT5 is the enzyme that methylates Aub. Our findings underscore the significance of sDMA modification of Piwi proteins in the germline and suggest an interacting pathway of genes that are required for piRNA function and PGC specification.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Kim, V. N. Small RNAs just got bigger: Piwi-interacting RNAs (piRNAs) in mammalian testes. Genes Dev. 20, 1993–1997 (2006).
O'Donnell K, A. & Boeke, J. D. Mighty Piwis Defend the Germline against Genome Intruders. Cell 129, 37–44 (2007).
Hartig, J. V., Tomari, Y. & Forstemann, K. piRNAs--the ancient hunters of genome invaders. Genes Dev. 21, 1707–1713 (2007).
Harris, A. N. & Macdonald, P. M. Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development 128, 2823–32 (2001).
Anne, J., Ollo, R., Ephrussi, A. & Mechler, B. M. Arginine methyltransferase Capsuleen is essential for methylation of spliceosomal Sm proteins and germ cell formation in Drosophila. Development 134, 137–146 (2007).
Gonsalvez, G. B., Rajendra, T. K., Tian, L. & Matera, A. G. The Sm-protein methyltransferase, dart5, is essential for germ-cell specification and maintenance. Curr. Biol. 16, 1077–1089 (2006).
Cox, D. N. et al. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 12, 3715–3727 (1998).
Gunawardane, L. S. et al. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315, 1587–1590 (2007).
Brennecke, J. et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128, 1089–1103 (2007).
Girard, A. & Hannon, G. J. Conserved themes in small-RNA-mediated transposon control. Trends Cell Biol. 18, 136–148 (2008).
Aravin, A. A. et al. The small RNA profile during Drosophila melanogaster development. Dev. Cell 5, 337–350 (2003).
Sarot, E., Payen-Groschene, G., Bucheton, A. & Pelisson, A. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics 166, 1313–1321 (2004).
Siomi, M. C., Saito, K. & Siomi, H. How selfish retrotransposons are silenced in Drosophila germline and somatic cells. FEBS Lett. 582, 2473–2478 (2008).
Aravin, A. A., Sachidanandam, R., Girard, A., Fejes-Toth, K. & Hannon, G. J. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316, 744–747 (2007).
Houwing, S. et al. A Role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell 129, 69–82 (2007).
Krause, C. D. et al. Protein arginine methyltransferases: evolution and assessment of their pharmacological and therapeutic potential. Pharmacol. Ther. 113, 50–87 (2007).
Bedford, M. T. & Richard, S. Arginine methylation an emerging regulator of protein function. Mol. Cell 18, 263–272 (2005).
Friesen, W. J. et al. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol. Cell Biol. 21, 8289–300 (2001).
Meister, G. et al. Methylation of Sm proteins by a complex containing PRMT5 and the putative U snRNP assembly factor pICln. Curr. Biol. 11, 1990–1994 (2001).
Lerner, E. A., Lerner, M. R., Janeway, C. A., Jr & Steitz, J. A. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc. Natl Acad. Sci. USA 78, 2737–2741 (1981).
Brahms, H. et al. The C-terminal RG dipeptide repeats of the spliceosomal Sm proteins D1 and D3 contain symmetrical dimethylarginines, which form a major B-cell epitope for anti-Sm autoantibodies. J. Biol. Chem. 275, 17122–17129 (2000).
Boisvert, F. M., Cote, J., Boulanger, M. C. & Richard, S. A proteomic analysis of arginine-methylated protein complexes. Mol. Cell Proteomics 2, 1319–1330 (2003).
Bowes, J. B. et al. Xenbase: a Xenopus biology and genomics resource. Nucleic Acids Res. 36, 761–767 (2008).
Kirino, Y. & Mourelatos, Z. Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nature Struct. Mol. Biol. 14, 347–348 (2007).
Ohara, T. et al. The 3′ termini of mouse Piwi-interacting RNAs are 2′-O-methylated. Nature Struct. Mol. Biol. 14, 349–350 (2007).
Saito, K. et al. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi- interacting RNAs at their 3′ ends. Genes Dev. 21, 1603–1608 (2007).
Horwich, M. D. et al. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr. Biol. 17, 1265–1272 (2007).
Gonsalvez, G. B. et al. Two distinct arginine methyltransferases are required for biogenesis of Sm-class ribonucleoproteins. J. Cell Biol. 178, 733–740 (2007).
Gonsalvez, G. B., Praveen, K., Hicks, A. J., Tian, L. & Matera, A. G. Sm protein methylation is dispensable for snRNP assembly in Drosophila melanogaster. RNA 14, 878–887 (2008).
Vagin, V. V. et al. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313, 320–324 (2006).
Cox, D. N., Chao, A. & Lin, H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development 127, 503–514 (2000).
Strome, S. & Lehmann, R. Germ versus soma decisions: lessons from flies and worms. Science 316, 392–393 (2007).
Anne, J. & Mechler, B. M. Valois, a component of the nuage and pole plasm, is involved in assembly of these structures, and binds to Tudor and the methyltransferase Capsuleen. Development 132, 2167–2177 (2005).
Boswell, R. E. & Mahowald, A. P. tudor, a gene required for assembly of the germ plasm in Drosophila melanogaster. Cell 43, 97–104 (1985).
Arkov, A. L., Wang, J. Y., Ramos, A. & Lehmann, R. The role of Tudor domains in germline development and polar granule architecture. Development 133, 4053–4062 (2006).
Selenko, P. et al. SMN tudor domain structure and its interaction with the Sm proteins. Nature Struct. Biol. 8, 27–31 (2001).
Cote, J. & Richard, S. Tudor domains bind symmetrical dimethylated arginines. J. Biol. Chem. 280, 28476–28483 (2005).
Chuma, S. et al. Tdrd1/Mtr-1, a tudor-related gene, is essential for male germ-cell differentiation and nuage/germinal granule formation in mice. Proc. Natl Acad. Sci. USA 103, 15894–15899 (2006).
Savitsky, M., Kwon, D., Georgiev, P., Kalmykova, A. & Gvozdev, V. Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Genes Dev. 20, 345–354 (2006).
Lim, A. K. & Kai, T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc. Natl Acad. Sci. USA 104, 6714–6719 (2007).
Acknowledgements
We are grateful to M.C. Siomi, H. Siomi, K. Saito, G. Dreyfuss for antibodies; to J. Anne for csul flies; to S. Kuramochi-Miyagawa and T. Nakano for Miwi and Mili cDNA constructs; to Rebecca Beerman for help with fly methodology; to Y. Kawamura for immunofluorescence protocols and to members of the Mourelatos lab for discussions. Mass spectrometry was performed at the Proteomics Core Facility (University of Pennsylvania) and at the Proteomics Resource of the Keck Foundation (Yale University). Illumina sequencing was performed at the Functional Genomics Core of the University of Pennsylvania. Protein production was at the Protein Expression Core of the Wistar Institute. We apologize to colleagues whose studies were not cited because of space limitations. This work was supported by a Human Frontier Science Program Long Term Fellowship to Y.K., and NIH grants to T.A.J. (NS046573), P.S.K. (GM76621) and Z.M.(GM0720777, NS056070, UL1RR024134). Z.M. also received ITMAT-PENN and URF-PENN.
Author information
Authors and Affiliations
Contributions
Y.K. and Z.M. conceived and designed the experiments; Y.K., N.K., M.P-S, E.K., S.C., P.S.K. and I.R. performed the experiments and analysis and generated all the figures; T.A.J. guided the Drosophila methodology and experiments; I.R. performed the bioinformatics analysis; T.A.J. and I.R. provided substantial input into the writing of the manuscript; Y.K. and Z.M. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 1116 kb)
Rights and permissions
About this article
Cite this article
Kirino, Y., Kim, N., de Planell-Saguer, M. et al. Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat Cell Biol 11, 652–658 (2009). https://doi.org/10.1038/ncb1872
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb1872
This article is cited by
-
Arginine methylation promotes siRNA-binding specificity for a spermatogenesis-specific isoform of the Argonaute protein CSR-1
Nature Communications (2021)
-
RNase κ promotes robust piRNA production by generating 2′,3′-cyclic phosphate-containing precursors
Nature Communications (2021)
-
Binding of guide piRNA triggers methylation of the unstructured N-terminal region of Aub leading to assembly of the piRNA amplification complex
Nature Communications (2021)
-
Comparative Proteomics Reveal Me31B’s Interactome Dynamics, Expression Regulation, and Assembly Mechanism into Germ Granules during Drosophila Germline Development
Scientific Reports (2020)
-
PRMT5 promotes DNA repair through methylation of 53BP1 and is regulated by Src-mediated phosphorylation
Communications Biology (2020)