Abstract
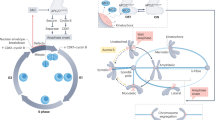
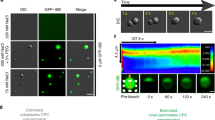
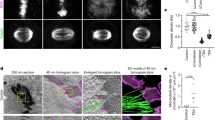
In Saccharomyces cerevisiae1 and HeLa cells2, the NoCut checkpoint, which involves the chromosome passenger kinase Aurora B, delays the completion of cytokinesis in response to anaphase defects. However, how NoCut monitors anaphase progression has not been clear. Here, we show that retention of chromatin in the plane of cleavage is sufficient to trigger NoCut, provided that Aurora/Ipl1 localizes properly to the spindle midzone, and that the ADA histone acetyltransferase complex is intact. Furthermore, forcing Aurora onto chromatin was sufficient to activate NoCut independently of anaphase defects. These findings provide the first evidence that NoCut is triggered by the interaction of acetylated chromatin with the passenger complex at the spindle midzone.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Norden, C. et al. The NoCut pathway links completion of cytokinesis to spindle midzone function to prevent chromosome breakage. Cell 125, 85–98 (2006).
Steigemann, P. et al. Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell 136, 473–484 (2009).
Musacchio, A. & Salmon, E. D. The spindle-assembly checkpoint in space and time. Nature Rev. Mol. Cell Biol. 8, 379–393 (2007).
May, K. M. & Hardwick, K. G. The spindle checkpoint. J. Cell Sci. 119, 4139–4142 (2006).
Barr, F. A. & Gruneberg, U. Cytokinesis: placing and making the final cut. Cell 131, 847–860 (2007).
Eggert, U. S., Mitchison, T. J. & Field, C. M. Animal cytokinesis: from parts list to mechanisms. Annu. Rev. Biochem. 75, 543–566 (2006).
Ruchaud, S., Carmena, M. & Earnshaw, W. C. Chromosomal passengers: conducting cell division. Nature Rev. Mol. Cell Biol. 8, 798–812 (2007).
Vader, G., Medema, R. H. & Lens, S. M. The chromosomal passenger complex: guiding Aurora-B through mitosis. J. Cell Biol. 173, 833–837 (2006).
Holm, C., Goto, T., Wang, J. C. & Botstein, D. DNA topoisomerase II is required at the time of mitosis in yeast. Cell 41, 553–563 (1985).
Uhlmann, F., Lottspeich, F. & Nasmyth, K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature 400, 37–42 (1999).
Melo, J. A., Cohen, J. & Toczyski, D. P. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 15, 2809–2821 (2001).
Uhlmann, F., Wernic, D., Poupart, M. A., Koonin, E. V. & Nasmyth, K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell 103, 375–386 (2000).
Queralt, E., Lehane, C., Novak, B. & Uhlmann, F. Downregulation of PP2A(Cdc55) phosphatase by separase initiates mitotic exit in budding yeast. Cell 125, 719–732 (2006).
Stegmeier, F., Visintin, R. & Amon, A. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell 108, 207–220 (2002).
Sullivan, M. & Uhlmann, F. A non-proteolytic function of separase links the onset of anaphase to mitotic exit. Nature Cell Biol. 5, 249–254 (2003).
Stegmeier, F. & Amon, A. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu. Rev. Genet. 38, 203–232 (2004).
Pereira, G. & Schiebel, E. Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14. Science 302, 2120–2124 (2003).
Higuchi, T. & Uhlmann, F. Stabilization of microtubule dynamics at anaphase onset promotes chromosome segregation. Nature 433, 171–176 (2005).
Eberharter, A. et al. The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 6621–6631 (1999).
Kelly, A. E. et al. Chromosomal enrichment and activation of the aurora B pathway are coupled to spatially regulate spindle assembly. Dev. Cell 12, 31–43 (2007).
King, E. M., Rachidi, N., Morrice, N., Hardwick, K. G. & Stark, M. J. Ipl1p-dependent phosphorylation of Mad3p is required for the spindle checkpoint response to lack of tension at kinetochores. Genes Dev. 21, 1163–1168 (2007).
Ramos, J. L. et al. The TetR family of transcriptional repressors. Microbiol Mol. Biol. Rev. 69, 326–356 (2005).
Alvaro, D., Lisby, M. & Rothstein, R. Genome-wide analysis of Rad52 foci reveals diverse mechanisms impacting recombination. PLoS Genet. 3, e228 (2007).
Yanagida, M. Fission yeast cut mutations revisited: control of anaphase. Trends Cell Biol. 8, 144–149 (1998).
Baxter, J. & Diffley, J. F. Topoisomerase II inactivation prevents the completion of DNA replication in budding yeast. Mol. Cell 30, 790–802 (2008).
Huh, W. K. et al. Global analysis of protein localization in budding yeast. Nature 425, 686–691 (2003).
Winzeler, E. A. et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906 (1999).
Sikorski, R. S. & Hieter, P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27 (1989).
Christianson, T. W., Sikorski, R. S., Dante, M., Shero, J. H. & Hieter, P. Multifunctional yeast high-copy-number shuttle vectors. Gene 110, 119–122 (1992).
Kusch, J., Meyer, A., Snyder, M. P. & Barral, Y. Microtubule capture by the cleavage apparatus is required for proper spindle positioning in yeast. Genes Dev. 16, 1627–1639 (2002).
Acknowledgements
We are grateful to Patrick Steigemann, Daniel Gerlich, Patrick Meraldi, Hemmo Meyer and all members of the Barral lab for fruitful discussions and critical reading of the manuscript. Thanks to Dominik Theler, Trinidad Sanmartin and Joelle Sasse for technical assistance, Orna Cohen-Fix for sharing reagents, and the ETH Light Microscopy Center for their invaluable support. This work was supported by an SNF Grant to Y.B. (2-77542-04).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 496 kb)
Rights and permissions
About this article
Cite this article
Mendoza, M., Norden, C., Durrer, K. et al. A mechanism for chromosome segregation sensing by the NoCut checkpoint. Nat Cell Biol 11, 477–483 (2009). https://doi.org/10.1038/ncb1855
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb1855
This article is cited by
-
The many functions of ESCRTs
Nature Reviews Molecular Cell Biology (2020)
-
Roles of the PH, coiled-coil and SAM domains of the yeast polarity protein Boi2 in polarity-site localization and function in polarized growth
Current Genetics (2020)
-
Building bridges between chromosomes: novel insights into the abscission checkpoint
Cellular and Molecular Life Sciences (2019)
-
Chromosomal instability induced by increased BIRC5/Survivin levels affects tumorigenicity of glioma cells
BMC Cancer (2017)
-
Oxidation of F-actin controls the terminal steps of cytokinesis
Nature Communications (2017)