Abstract
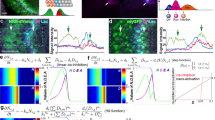
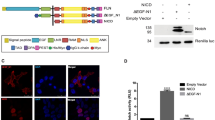
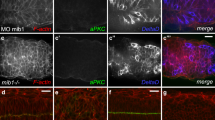
In canonical Delta–Notch signalling, expression of Delta activates Notch in neighbouring cells, leading to downregulation of Delta in these cells1. This process of lateral inhibition results in selection of either Delta-signalling cells or Notch-signalling cells. Here we show that d-Asb11 is an important mediator of this lateral inhibition. In zebrafish embryos, morpholino oligonucleotide (MO)-mediated knockdown of d-Asb11 caused repression of specific Delta–Notch elements and their transcriptional targets, whereas these were induced when d-Asb11 was misexpressed. d-Asb11 also activated legitimate Notch reporters cell-non-autonomously in vitro and in vivo when co-expressed with a Notch reporter. However, it repressed Notch reporters when expressed in Delta-expressing cells. Consistent with these results, d-Asb11 was able to specifically ubiquitylate and degrade DeltaA both in vitro and in vivo. We conclude that d-Asb11 is a component in the regulation of Delta–Notch signalling, important in fine-tuning the lateral inhibition gradients between DeltaA and Notch through a cell non-autonomous mechanism.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Louvi, A. & Artavanis-Tsakonas, S. Notch signalling in vertebrate neural development. Nature Rev. Neurosci. 7, 93–102 (2006).
Lai, E. C. Notch signaling: control of cell communication and cell fate. Development 131, 965–973 (2004).
Mumm, J. S. & Kopan, R. Notch signaling: from the outside in. Dev. Biol. 228, 151–165 (2000).
Crowe, R., Henrique, D., Ish-Horowicz, D., & Niswander, L. A new role for Notch and Delta in cell fate decisions: patterning the feather array. Development 125, 767–775 (1998).
Kopan, R. & Turner, D. L. The Notch pathway: democracy and aristocracy in the selection of cell fate. Curr. Opin. Neurobiol. 6, 594–601 (1996).
Le Borgne, R. Regulation of Notch signalling by endocytosis and endosomal sorting. Curr. Opin. Cell Biol. 18, 213–222 (2006).
Chitnis, A. Why is delta endocytosis required for effective activation of notch? Dev. Dyn. 235, 886–894 (2006).
Baron, M. et al. Multiple levels of Notch signal regulation (review). Mol. Membr. Biol. 19, 27–38 (2002).
Wilkin, M. B. et al. Regulation of notch endosomal sorting and signaling by Drosophila Nedd4 family proteins. Curr. Biol. 14, 2237–2244 (2004).
Gupta-Rossi, N. et al. Monoubiquitination and endocytosis direct γ-secretase cleavage of activated Notch receptor. J. Cell Biol. 166, 73–83 (2004).
Wu, G. et al. SEL-10 is an inhibitor of notch signaling that targets notch for ubiquitin-mediated protein degradation. Mol. Cell Biol. 21, 7403–7415 (2001).
Itoh, M. et al. Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev. Cell 4, 67–82 (2003).
Lai, E. C., Deblandre, G. A., Kintner, C., & Rubin, G. M. Drosophila neuralized is a ubiquitin ligase that promotes the internalization and degradation of delta. Dev. Cell 1, 783–794 (2001).
Pitsouli, C. & Delidakis, C. The interplay between DSL proteins and ubiquitin ligases in Notch signaling. Development 132, 4041–4050 (2005).
Wang, W. & Struhl, G. Distinct roles for Mind bomb, Neuralized and Epsin in mediating DSL endocytosis and signaling in Drosophila. Development 132, 2883–2894 (2005).
Diks, S. H. et al. The novel gene asb11: a regulator of the size of the neural progenitor compartment. J. Cell Biol. 174, 581–592 (2006).
Appel, B. & Eisen, J. S. Regulation of neuronal specification in the zebrafish spinal cord by Delta function. Development 125, 371–380 (1998).
Jarriault, S. et al. Signalling downstream of activated mammalian Notch. Nature 377, 355–358 (1995).
Nishimura, M. et al. Structure, chromosomal locus, and promoter of mouse Hes2 gene, a homologue of Drosophila hairy and Enhancer of split. Genomics 49, 69–75 (1998).
Krebs, L. T., Deftos, M. L., Bevan, M. J., & Gridley, T. The Nrarp gene encodes an ankyrin-repeat protein that is transcriptionally regulated by the notch signaling pathway. Dev. Biol. 238, 110–119 (2001).
Lamar, E. et al. Nrarp is a novel intracellular component of the Notch signaling pathway. Genes Dev. 15, 1885–1899 (2001).
Ishitani, T., Matsumoto, K., Chitnis, A. B., & Itoh, M. Nrarp functions to modulate neural-crest-cell differentiation by regulating LEF1 protein stability. Nature Cell Biol. 7, 1106–1112 (2005).
Deblandre, G. A., Lai, E. C., & Kintner, C. Xenopus neuralized is a ubiquitin ligase that interacts with XDelta1 and regulates Notch signaling. Dev. Cell 1, 795–806 (2001).
Chitnis, A., Henrique, D., Lewis, J., Ishhorowicz, D., & Kintner, C. Primary neurogenesis in Xenopus embryos regulated by a homolog of the Drosophila neurogenic gene-delta. Nature 375, 761–766 (1995).
Bijlsma, M. F. et al. Repression of smoothened by patched-dependent (Pro-) vitamin D3 secretion. PLoS. Biol. 4, e232 (2006).
Yeo, S. Y., Kim, M., Kim, H. S., Huh, T. L., & Chitnis, A. B. Fluorescent protein expression driven by her4 regulatory elements reveals the spatiotemporal pattern of Notch signaling in the nervous system of zebrafish embryos. Dev. Biol. 301, 555–567 (2007).
Schmitt, T. M. & Zuniga-Pflucker, J. C. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 17, 749–756 (2002).
Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B., & Schilling, T. F. Stages of embryonic development of the zebrafish. Dev. Dynam. 203, 253–310 (1995).
Westerfield, M. The Zebrafish Book (Eugene: University of Oregon Press, 1994).
Holley, S. A., Julich, D., Rauch, G. J., Geisler, R., & Nusslein-Volhard, C. her1 and the notch pathway function within the oscillator mechanism that regulates zebrafish somitogenesis. Development 129, 1175–1183 (2002).
Latimer, A. J., Dong, X. H., Markov, Y., & Appel, B. Delta-Notch signaling induces hypochord development in zebrafish. Development 129, 2555–2563 (2002).
Joore, J. et al. Effects of retinoic acid on the expression of retinoic acid receptors during zebrafish embryogenesis. Mech. Dev. 46, 137–150 (1994).
Cunliffe, V. T. & Casaccia-Bonnefil, P. Histone deacetylase 1 is essential for oligodendrocyte specification in the zebrafish CNS. Mech. Dev. 123, 24–30 (2006).
Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 29, e45 (2001).
Acknowledgements
We wish to thank J. A. Campos-Ortega for providing the MT–notch1a ICD construct, B. Appel for the MT–deltaA construct and A. Israel for the different Hes1–luciferase reporter constructs. We thank R. Dorsky, C. Houart, P. Ingham, J.P. Concordet for the riboprobes. We thank J. Lewis for DeltaD antibody, G. Strous for HA–Ubiquitin, U. Strähle for XDeltastu. We are indebted to R. Dorsky and R. Moon for providing TOPdGFP fish. We thank Y.J. Jiang for advice. We also wish to thank K. Österreicher and D. Kolmer (TissueGnostics, GmbH) for their help in the quantification of the co-localization studies described in Supplementary Information, Fig. S2. NWO Casimir, NWO Genomics, NWO ALW, TIPharma and the IAG program of the province of Groningen and the European Commission are thanked for financial support. We are especially grateful to S. van de Water, S. van den Brink, A. Visser and L. Glazenburg for technical assistance and to the animal facility of the Hubrecht Institute for care of zebrafish.
Author information
Authors and Affiliations
Contributions
S.H.D. contributed substantially to the conception and design of the study, acquisition, analysis and interpretation of the data, and drafting the manuscript; M.A.S.d.S. acquired, analysed and interpreted data; J.L.H., R.J.B., H.H.V., C.v.R. and A.B. acquired, analysed and interpreted data, and provided critical comments; M.P.P. analysed and interpreted the data; D.Z. acquired, analysed and interpreted data; M.P.P. and D.Z conceived and designed the study, and wrote and revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures S1, S2, S3, S4, S5, Supplementary Data and Supplementary Table 1 (PDF 805 kb)
Rights and permissions
About this article
Cite this article
Diks, S., Sartori da Silva, M., Hillebrands, JL. et al. d-Asb11 is an essential mediator of canonical Delta–Notch signalling. Nat Cell Biol 10, 1190–1198 (2008). https://doi.org/10.1038/ncb1779
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb1779
This article is cited by
-
The role of cullin 5-containing ubiquitin ligases
Cell Division (2016)