Abstract
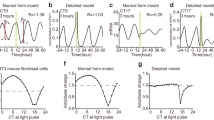
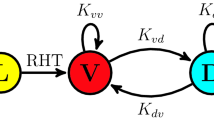
Singularity behaviour in circadian clocks1,2 — the loss of robust circadian rhythms following exposure to a stimulus such as a pulse of bright light — is one of the fundamental but mysterious properties of clocks. To quantitatively perturb and accurately measure the dynamics of cellular clocks3,4, we synthetically produced photo-responsiveness within mammalian cells by exogenously introducing the photoreceptor melanopsin5,6,7,8 and continuously monitoring the effect of photo-perturbation on the state of cellular clocks. Here we report that a critical light pulse drives cellular clocks into singularity behaviour. Our theoretical analysis consistently predicts and subsequent single-cell level observation directly proves that desynchronization of individual cellular clocks underlies singularity behaviour. Our theoretical framework also explains why singularity behaviours have been experimentally observed in various organisms, and it suggests that desynchronization is a plausible mechanism for the observable singularity of circadian clocks. Importantly, these in vitro and in silico findings are further supported by in vivo observations that desynchronization underlies the multicell-level amplitude decrease in the rat suprachiasmatic nucleus induced by critical light pulses.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Winfree, A. T. Unclocklike behaviour of biological clocks. Nature 253, 315–319 (1975).
Winfree, A. T. The Geometry of Biological Time (Springer-Verlag, New York, 1980).
Balsalobre, A., Damiola, F. & Schibler, U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93, 929–937 (1998).
Akashi, M. & Nishida, E. Involvement of the MAP kinase cascade in resetting of the mammalian circadian clock. Genes Dev. 14, 645–649 (2000).
Melyan, Z., Tarttelin, E. E., Bellingham, J., Lucas, R. J. & Hankins, M. W. Addition of human melanopsin renders mammalian cells photoresponsive. Nature 433, 741–745 (2005).
Qiu, X. et al. Induction of photosensitivity by heterologous expression of melanopsin. Nature 433, 745–749 (2005).
Panda, S. et al. Illumination of the melanopsin signaling pathway. Science 307, 600–604 (2005).
Peirson, S. & Foster, R. G. Melanopsin: another way of signaling light. Neuron 49, 331–339 (2006).
Ueda, H. R. et al. A transcription factor response element for gene expression during circadian night. Nature 418, 534–539 (2002).
Ueda, H. R. et al. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nature Genet. 37, 187–192 (2005).
Sato, T. K. et al. Feedback repression is required for mammalian circadian clock function. Nature Genet. 38, 312–319 (2006).
Johnson, C. H., Elliott, J., Foster, R., Honma, K. & Kronauer, R. in Chronobiology: Biological Timekeeping (eds Dunlap, J. C., Loros, J. J. & DeCoursey, P. J.) 67–106 (Sinauer, Sunderland, Massachusetts, 2004).
Johnson, C. H. Forty years of PRCs–what have we learned? Chronobiol. Int. 16, 711–743 (1999).
Johnson, C.H. & Kondo, T. Light pulses induce 'singular' behavior and shorten the period of the circadian phototaxis rhythm in the CW15 strain of Chlamydomonas. J. Biol. Rhythms 7, 313–327 (1992).
Welsh, D. K., Yoo, S. H., Liu, A. C., Takahashi, J. S. & Kay, S. A. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr. Biol. 14, 2289–2295 (2004).
Nagoshi, E. et al. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell 119, 693–705 (2004).
Jewett, M. E., Kronauer, R. E. & Czeisler, C. A. Light-induced suppression of endogenous circadian amplitude in humans. Nature 350, 59–62 (1991).
Honma, S. & Honma, K. Light-induced uncoupling of multioscillatory circadian system in a diurnal rodent, Asian chipmunk. Am. J. Physiol. 276, R1390–R1396 (1999).
Maywood, E. S. et al. Synchronization and maintenance of timekeeping in suprachiasmatic circadian clock cells by neuropeptidergic signaling. Curr. Biol. 16, 599–605 (2006).
Yamaguchi, S. et al. Synchronization of cellular clocks in the suprachiasmatic nucleus. Science 302, 1408–1412 (2003).
Leloup, J. C., Gonze, D. & Goldbeter, A. Limit cycle models for circadian rhythms based on transcriptional regulation in Drosophila and Neurospora. J. Biol. Rhythms 14, 433–448 (1999).
Leloup, J. C. & Goldbeter, A. A molecular explanation for the long-term suppression of circadian rhythms by a single light pulse. Am. J. Physiol. Regul. Integr. Comp. Physiol. 280, R1206–R1212 (2001).
Huang, G., Wang, L. & Liu, Y. Molecular mechanism of suppression of circadian rhythms by a critical stimulus. EMBO J. (2006).
Yan, L., Takekida, S., Shigeyoshi, Y. & Okamura, H. Per1 and Per2 gene expression in the rat suprachiasmatic nucleus: circadian profile and the compartment-specific response to light. Neuroscience 94, 141–150 (1999).
Ban, Y., Shigeyoshi, Y. & Okamura, H. Development of vasoactive intestinal peptide mRNA rhythm in the rat suprachiasmatic nucleus. J. Neurosci. 17, 3920–3931 (1997).
Acknowledgements
This research was supported by intramural Grant-in-Aid from the Center for Developmental Biology (CDB), Director's Fund from CDB (H.R.U.), KAKENHI (Grant-in-Aid for Scientific Research) on Priority Areas 'Systems Genomics' from the Ministry of Education, Culture, Sports, Science and Technology of Japan (H.R.U. and H.U.), Grant-in-Aid for Scientific Research on 'Development of Basic Technology to Control Biological Systems Using Chemical Compounds' from the New Energy and Industrial Technology Development Organization (NEDO) of Japan (H.R.U.) and Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists 17-4169 (T.J.K). We thank for Wataru Kishimoto for technical supports on high-throughput monitoring system, Rikuhiro G. Yamada for support for data analysis, Maki Ukai-Tadenuma for technical support, Yoichi Minami for discussion for in vivo experiments, and Douglas Sipp Michael Royle, Danny Forger for critical comments on the manuscript. We also thank John J. Tyson for his suggestion regarding Winfree's original idea of desynchronization.
Author information
Authors and Affiliations
Contributions
K.Y. and H.R.U. developed the concept of synthetic implementation of photo-responsiveness within NIH3T3 cells by using melanopsin. T.K. designed a high-throughput monitoring device. H.U. constructed all materials used in this work and performed high-throughput real-time luciferase reporter assays, single-cell real-time bioluminescence imaging and pharmacological assays. T.J.K. developed the theory, data-analysis methods, and the image-analysis software used in this work, analysed high-throughput, single-cell real-time bioluminescence, in situ hybridization, and locomotor activity data, and performed single-cell real-time bioluminescence imaging. M.N. and M.S. performed behaviour analysis of rats. M.N., K.M. and Y.S. performed in situ hybridization and analysed the data. H.U., T.J.K. and H.R.U wrote the manuscript. All authors discussed the results and commented on the manuscript text.
Corresponding author
Supplementary information
Supplementary Information
Supplementary figures S1 to S11 and Supplementary Table S1 (PDF 2563 kb)
Supplementary Information
Supplementary Movie 1 (MOV 4034 kb)
Supplementary Information
Supplementary Movie 2 (MOV 846 kb)
Supplementary Information
Supplementary Movie 3 (MOV 5663 kb)
Rights and permissions
About this article
Cite this article
Ukai, H., Kobayashi, T., Nagano, M. et al. Melanopsin-dependent photo-perturbation reveals desynchronization underlying the singularity of mammalian circadian clocks. Nat Cell Biol 9, 1327–1334 (2007). https://doi.org/10.1038/ncb1653
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb1653
This article is cited by
-
Amplitude response and singularity behavior of circadian clock to external stimuli
npj Systems Biology and Applications (2023)
-
Singularity response reveals entrainment properties in mammalian circadian clock
Nature Communications (2023)
-
Regulation of molecular clock oscillations and phagocytic activity via muscarinic Ca2+ signaling in human retinal pigment epithelial cells
Scientific Reports (2017)
-
The mammalian circadian clock and its entrainment by stress and exercise
The Journal of Physiological Sciences (2017)
-
Rigid Cooperation of Per1 and Per2 proteins
Scientific Reports (2016)