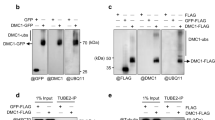
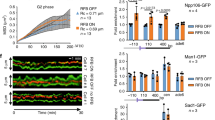
Abstract
Homologous recombination (HR) is crucial for maintaining genome integrity by repairing DNA double-strand breaks (DSBs) and rescuing collapsed replication forks. In contrast, uncontrolled HR can lead to chromosome translocations, loss of heterozygosity, and deletion of repetitive sequences. Controlled HR is particularly important for the preservation of repetitive sequences of the ribosomal gene (rDNA) cluster. Here we show that recombinational repair of a DSB in rDNA in Saccharomyces cerevisiae involves the transient relocalization of the lesion to associate with the recombination machinery at an extranucleolar site. The nucleolar exclusion of Rad52 recombination foci entails Mre11 and Smc5–Smc6 complexes and depends on Rad52 SUMO (small ubiquitin-related modifier) modification. Remarkably, mutations that abrogate these activities result in the formation of Rad52 foci within the nucleolus and cause rDNA hyperrecombination and the excision of extrachromosomal rDNA circles. Our study also suggests a key role of sumoylation for nucleolar dynamics, perhaps in the compartmentalization of nuclear activities.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Leger-Silvestre, I., Trumtel, S., Noaillac-Depeyre, J. & Gas, N. Functional compartmentalization of the nucleus in the budding yeast Saccharomyces cerevisiae. Chromosoma 108, 103–113 (1999).
Michel, A. H., Kornmann, B., Dubrana, K. & Shore, D. Spontaneous rDNA copy number variation modulates Sir2 levels and epigenetic gene silencing. Genes Dev. 19, 1199–1210 (2005).
Park, P. U., Defossez, P. A. & Guarente, L. Effects of mutations in DNA repair genes on formation of ribosomal DNA circles and life span in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 3848–3856 (1999).
Kobayashi, T., Heck, D. J., Nomura, M. & Horiuchi, T. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 12, 3821–3830 (1998).
Gangloff, S., Zou, H. & Rothstein, R. Gene conversion plays the major role in controlling the stability of large tandem repeats in yeast. EMBO J. 15, 1715–1725 (1996).
Warner, J. R., Vilardell, J. & Sohn, J. H. Economics of ribosome biosynthesis. Cold Spring Harb. Symp. Quant. Biol. 66, 567–574 (2001).
Liu, Y., Li, M., Lee, E. Y. & Maizels, N. Localization and dynamic relocalization of mammalian Rad52 during the cell cycle and in response to DNA damage. Curr. Biol. 9, 975–978 (1999).
Lisby, M., Rothstein, R. & Mortensen, U. H. Rad52 forms DNA repair and recombination centers during S phase. Proc. Natl Acad. Sci. USA 98, 8276–8282 (2001).
Schimmang, T., Tollervey, D., Kern, H., Frank, R. & Hurt, E. C. A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. EMBO J. 8, 4015–4024 (1989).
Lisby, M., Mortensen, U. H. & Rothstein, R. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nature Cell Biol. 5, 572–577 (2003).
Lisby, M., Barlow, J. H., Burgess, R. C. & Rothstein, R. Choreography of the DNA damage response; spatiotemporal relationships among checkpoint and repair proteins. Cell 118, 699–713 (2004).
de Jager, M. et al. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol. Cell 8, 1129–1135 (2001).
Krogh, B. & Symington, L. Recombination proteins in yeast. Annu. Rev. Genet. 38, 233–271 (2004).
Zou, L. & Elledge, S. J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300, 1542–1548 (2003).
Torres-Rosell, J. et al. Anaphase onset before complete DNA replication with intact checkpoint responses. Science 315, 1411–1415 (2007).
Fujioka, Y., Kimata, Y., Nomaguchi, K., Watanabe, K. & Kohno, K. Identification of a novel non-structural maintenance of chromosomes (SMC) component of the SMC5–SMC6 complex involved in DNA repair. J. Biol. Chem. 277, 21585–21591 (2002).
Zhao, X. & Blobel, G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl Acad. Sci. USA 102, 4777–4782 (2005).
Losada, A. & Hirano, T. Dynamic molecular linkers of the genome: the first decade of SMC proteins. Genes Dev. 19, 1269–1287 (2005).
Aragon, L. Sumoylation: a new wrestler in the DNA repair ring. Proc. Natl Acad. Sci. USA 102, 4661–4662 (2005).
Torres-Rosell, J. et al. SMC5 and SMC6 genes are required for the segregation of repetitive chromosome regions. Nature Cell Biol. 7, 412–419 (2005).
Kobayashi, T., Horiuchi, T., Tongaonkar, P., Vu, L. & Nomura, M. SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell 117, 441–453 (2004).
Ivessa, A. S., Zhou, J. Q. & Zakian, V. A. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell 100, 479–489 (2000).
Eladad, S. et al. Intra-nuclear trafficking of the BLM helicase to DNA damage-induced foci is regulated by SUMO modification. Hum. Mol. Genet. 14, 1351–1365 (2005).
Shen, T. H., Lin, H. K., Scaglioni, P. P., Yung, T. M. & Pandolfi, P. P. The mechanisms of PML-nuclear body formation. Mol. Cell 24, 331–339 (2006).
Lin, D. Y. et al. Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol. Cell 24, 341–354 (2006).
Sacher, M., Pfander, B., Hoege, C. & Jentsch, S. Control of Rad52 recombination activity by double-strand break-induced SUMO modification. Nature Cell Biol. 8, 1284–1290 (2006).
De Piccoli, G. et al. Smc5–Smc6 mediate DNA double-strand-break repair by promoting sister-chromatid recombination. Nature Cell Biol. 8, 1032–1034 (2006).
Betts Lindroos, H. et al. Chromosomal association of the Smc5/6 complex reveals that it functions in differently regulated pathways. Mol. Cell 22, 755–767 (2006).
Veaute, X. et al. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423, 309–312 (2003).
Torres, J. Z., Schnakenberg, S. L. & Zakian, V. A. Saccharomyces cerevisiae Rrm3p DNA helicase promotes genome integrity by preventing replication fork stalling: viability of rrm3 cells requires the intra-S-phase checkpoint and fork restart activities. Mol. Cell. Biol. 24, 3198–3212 (2004).
Hays, S. L., Firmenich, A. A. & Berg, P. Complex formation in yeast double-strand break repair: participation of Rad51, Rad52, Rad55, and Rad57 proteins. Proc. Natl Acad. Sci. USA 92, 6925–6929 (1995).
Davis, A. P. & Symington, L. S. The yeast recombinational repair protein Rad59 interacts with Rad52 and stimulates single-strand annealing. Genetics 159, 515–525 (2001).
Ho, J. C., Warr, N. J., Shimizu, H. & Watts, F. Z. SUMO modification of Rad22, the Schizosaccharomyces pombe homologue of the recombination protein Rad52. Nucleic Acids Res. 29, 4179–4186 (2001).
Reid, R., Lisby, M. & Rothstein, R. Cloning-free genome alterations in Saccharomyces cerevisiae using adaptamer-mediated PCR. Methods Enzymol. 350, 258–277 (2002).
Antúnez de Mayolo, A. et al. Multiple start codons and phosphorylation result in discrete Rad52 protein species. Nucleic Acids Res. 34, 2587–2597 (2006).
Erdeniz, N., Mortensen, U. H. & Rothstein, R. Cloning-free PCR-based allele replacement methods. Genome Res. 7, 1174–1183 (1997).
Merker, R. J. & Klein, H. L. hpr1Δ affects ribosomal DNA recombination and cell life span in Saccharomyces cerevisiae. Mol. Cell. Biol. 22, 421–429 (2002).
Versini, G. et al. The yeast Sgs1 helicase is differentially required for genomic and ribosomal DNA replication. EMBO J. 22, 1939–1949 (2003).
Moore, C. W. et al. DNA damage-inducible and RAD52-independent repair of DNA double-strand breaks in Saccharomyces cerevisiae. Genetics 154, 1085–1099 (2000).
Acknowledgements
We thank M. Wagner, R. Tsien, M. Jasin and K. Nasmyth for plasmids and strains, and members of the Rothstein, Aragon, Jentsch and Lisby laboratories for helpful discussions on this work. This work was supported by grants to R.R. from the NIH (GM50237 and GM67055), the Human Frontiers Science Program (RGP0238/2001) and the Tonnesen Foundation (RGP0238/2001), to M.L. from the Danish Natural Science Research Council, the Alfred Benzon Foundation, the Villum Kann Rasmussen Foundation, and to N.E.B. from the Lundbeck Foundation. The Aragon laboratory was supported by the Medical Research Council, UK.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figure S1, S2, S3, S4, S5, Table S1 and Supplementary Methods (PDF 706 kb)
Supplementary Information
Supplementary Movie 1 (MOV 215 kb)
Rights and permissions
About this article
Cite this article
Torres-Rosell, J., Sunjevaric, I., De Piccoli, G. et al. The Smc5–Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat Cell Biol 9, 923–931 (2007). https://doi.org/10.1038/ncb1619
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb1619
This article is cited by
-
The SAGA histone acetyltransferase module targets SMC5/6 to specific genes
Epigenetics & Chromatin (2023)
-
Nucleolar release of rDNA repeats for repair involves SUMO-mediated untethering by the Cdc48/p97 segregase
Nature Communications (2021)
-
DNA repair by Rad52 liquid droplets
Nature Communications (2020)
-
Biomolecular condensates as arbiters of biochemical reactions inside the nucleus
Communications Biology (2020)
-
The nuclear pore primes recombination-dependent DNA synthesis at arrested forks by promoting SUMO removal
Nature Communications (2020)