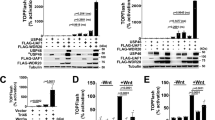
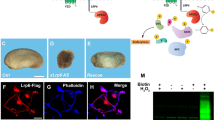
Abstract
Dishevelled is a conserved protein that interprets signals received by Frizzled receptors. Using a tandem-affinity purification strategy and mass spectrometry we have identified proteins associated with Dishevelled, including a Cullin-3 ubiquitin ligase complex containing the Broad Complex, Tramtrack and Bric à Brac (BTB) protein Kelch-like 12 (KLHL12). This E3 ubiquitin ligase complex is recruited to Dishevelled in a Wnt-dependent manner that promotes its poly-ubiquitination and degradation. Functional analyses demonstrate that regulation of Dishevelled by this ubiquitin ligase antagonizes the Wnt–ß-catenin pathway in cultured cells, as well as in Xenopus and zebrafish embryos. Considered with evidence that the distinct Cullin-1 based SCFβ-TrCPcomplex regulates β-catenin stability, our data on the stability of Dishevelled demonstrates that two distinct ubiquitin ligase complexes regulate the Wnt–ß-catenin pathway.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Latres, E., Chiaur, D. S. & Pagano, M. The human F box protein β-Trcp associates with the Cul1–Skp1 complex and regulates the stability of β-catenin. Oncogene 18, 849–854 (1999).
Hart, M. et al. The F-box protein β-TrCP associates with phosphorylated β-catenin and regulates its activity in the cell. Curr. Biol. 9, 207–210 (1999).
Wodarz, A. & Nusse, R. Mechanisms of Wnt signaling in development. Annu. Rev. Cell Dev. Biol. 14, 59–88 (1998).
Wharton, K. A., Jr. Runnin' with the Dvl: proteins that associate with Dsh–Dvl and their significance to Wnt signal transduction. Dev. Biol. 253, 1–17 (2003).
Wallingford, J. B. et al. Dishevelled controls cell polarity during Xenopus gastrulation. Nature 405, 81–85 (2000).
Mazieres, J., He, B., You, L., Xu, Z. & Jablons, D. M. Wnt signaling in lung cancer. Cancer Lett. 222, 1–10 (2005).
Hershko, A. & Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 (1998).
Simons, M. et al. Inversin, the gene product mutated in nephronophthisis type II, functions as a molecular switch between Wnt signaling pathways. Nature Genet. 37, 537–543 (2005).
Miyazaki, K. et al. NEDL1, a novel ubiquitin-protein isopeptide ligase for dishevelled-1, targets mutant superoxide dismutase-1. J. Biol. Chem. 279, 11327–11335 (2004).
Willert, K., Brink, M., Wodarz, A., Varmus, H. & Nusse, R. Casein kinase 2 associates with and phosphorylates dishevelled. EMBO J. 16, 3089–3096 (1997).
Cong, F., Schweizer, L. & Varmus, H. Casein kinase Iε modulates the signaling specificities of dishevelled. Mol. Cell Biol. 24, 2000–2011 (2004).
Sun, T. Q. et al. PAR-1 is a Dishevelled-associated kinase and a positive regulator of Wnt signalling. Nature Cell Biol. 3, 628–636 (2001).
Yan, D. et al. Cell autonomous regulation of multiple Dishevelled-dependent pathways by mammalian Nkd. Proc. Natl Acad. Sci. USA 98, 3802–3807 (2001).
Ratcliffe, M. J., Itoh, K. & Sokol, S. Y. A positive role for the PP2A catalytic subunit in Wnt signal transduction. J. Biol. Chem. 275, 35680–35683 (2000).
Park, M. & Moon, R. T. The planar cell-polarity gene stbm regulates cell behaviour and cell fate in vertebrate embryos. Nature Cell Biol. 4, 20–25 (2002).
Zhang, D. D. et al. Ubiquitination of Keap1, a BTB–Kelch substrate adaptor protein for Cul3, targets Keap1 for degradation by a proteasome-independent pathway. J. Biol. Chem. 280, 30091–30099 (2005).
Bomont, P. et al. The gene encoding gigaxonin, a new member of the cytoskeletal BTB–kelch repeat family, is mutated in giant axonal neuropathy. Nature Genet. 26, 370–374 (2000).
Petroski, M. D. & Deshaies, R. J. Function and regulation of cullin–RING ubiquitin ligases. Nature Rev. Mol. Cell Biol. 6, 9–20 (2005).
van den Heuvel, S. Protein degradation: CUL-3 and BTB — partners in proteolysis. Curr. Biol. 14, R59–R61 (2004).
Furukawa, M., He, Y. J., Borchers, C. & Xiong, Y. Targeting of protein ubiquitination by BTB–Cullin 3–Roc1 ubiquitin ligases. Nature Cell Biol. 5, 1001–1007 (2003).
Kelly, G. M., Greenstein, P., Erezyilmaz, D. F. & Moon, R. T. Zebrafish wnt8 and wnt8b share a common activity but are involved in distinct developmental pathways. Development 121, 1787–1799 (1995).
Dorsky, R. I., Sheldahl, L. C. & Moon, R. T. A transgenic Lef1–β-catenin-dependent reporter is expressed in spatially restricted domains throughout zebrafish development. Dev. Biol. 241, 229–237 (2002).
Sokol, S. Y. Analysis of Dishevelled signalling pathways during Xenopus development. Curr. Biol. 6, 1456–1467 (1996).
Sheldahl, L. C. et al. Dishevelled activates Ca2+ flux, PKC, and CamKII in vertebrate embryos. J. Cell Biol. 161, 769–777 (2003).
Torres, M. A. et al. Activities of the Wnt-1 class of secreted signaling factors are antagonized by the Wnt-5A class and by a dominant negative cadherin in early Xenopus development. J. Cell Biol. 133, 1123–1137 (1996).
Rothbacher, U. et al. Dishevelled phosphorylation, subcellular localization and multimerization regulate its role in early embryogenesis. EMBO J. 19, 1010–1022 (2000).
Katanaev, V. L., Ponzielli, R., Semeriva, M. & Tomlinson, A. Trimeric G protein-dependent frizzled signaling in Drosophila. Cell 120, 111–122 (2005).
Liu, X., Rubin, J. S. & Kimmel, A. R. Rapid, Wnt-induced changes in GSK3β associations that regulate β-catenin stabilization are mediated by Gα proteins. Curr. Biol. 15, 1989–1997 (2005).
Cardozo, T. & Pagano, M. The SCF ubiquitin ligase: insights into a molecular machine. Nature Rev. Mol. Cell Biol. 5, 739–751 (2004).
Willems, A. R., Schwab, M. & Tyers, M. A hitchhiker's guide to the cullin ubiquitin ligases: SCF and its kin. Biochim. Biophys. Acta 1695, 133–170 (2004).
Geyer, R., Wee, S., Anderson, S., Yates, J. & Wolf, D. A. BTB–POZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol. Cell 12, 783–790 (2003).
Krek, W. BTB proteins as henchmen of Cul3-based ubiquitin ligases. Nature Cell Biol. 5, 950–951 (2003).
Pintard, L. et al. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature 425, 311–316 (2003).
Xu, L. et al. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature 425, 316–321 (2003).
Cullinan, S. B., Gordan, J. D., Jin, J., Harper, J. W. & Diehl, J. A. The Keap1–BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol. Cell Biol. 24, 8477–8486 (2004).
Furukawa, M. & Xiong, Y. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3–Roc1 ligase. Mol. Cell Biol. 25, 162–171 (2005).
Kobayashi, A. et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell Biol. 24, 7130–7139 (2004).
Zhang, D. D., Lo, S. C., Cross, J. V., Templeton, D. J. & Hannink, M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell Biol. 24, 10941–10953 (2004).
Hernandez-Munoz, I. et al. Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3–SPOP ubiquitin E3 ligase. Proc. Natl Acad. Sci. USA 102, 7635–7640 (2005).
Yamamoto, H. et al. Phosphorylation of axin, a Wnt signal negative regulator, by glycogen synthase kinase-3β regulates its stability. J. Biol. Chem. 274, 10681–10684 (1999).
Choi, J., Park, S. Y., Costantini, F., Jho, E. H. & Joo, C. K. Adenomatous polyposis coli is down-regulated by the ubiquitin-proteasome pathway in a process facilitated by Axin. J. Biol. Chem. 279, 49188–49198 (2004).
Creyghton, M. P. et al. PR72, a novel regulator of Wnt signaling required for Naked cuticle function. Genes Dev. 19, 376–386 (2005).
Uematsu, K. et al. Activation of the Wnt pathway in non small cell lung cancer: evidence of dishevelled overexpression. Oncogene 22, 7218–7221 (2003).
Kaykas, A. et al. Mutant Frizzled 4 associated with vitreoretinopathy traps wild-type Frizzled in the endoplasmic reticulum by oligomerization. Nature Cell Biol. 6, 52–58 (2004).
Gingras, A. C. et al. A novel, evolutionarily conserved protein phosphatase complex involved in cisplatin sensitivity. Mol. Cell Proteomics 4, 1725–1740 (2005).
Gatlin, C. L., Kleemann, G. R., Hays, L. G., Link, A. J. & Yates, J. R., 3rd. Protein identification at the low femtomole level from silver-stained gels using a new fritless electrospray interface for liquid chromatography-microspray and nanospray mass spectrometry. Anal. Biochem. 263, 93–101 (1998).
MacCoss, M. J., Wu, C. C. & Yates, J. R., 3rd. Probability-based validation of protein identifications using a modified SEQUEST algorithm. Anal. Chem. 74, 5593–5599 (2002).
Tabb, D. L., McDonald, W. H. & Yates, J. R., 3rd. DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1, 21–26 (2002).
Acknowledgements
We thank A.-C. Gingras for insightful technical discussions. S.A. is an associate and R.T.M. an investigator of the Howard Hughes Medical Institute. M.M. was supported by National Institutes of Health-National Center for Research Resources (NIH-NCRR) grant (P41 RR011823).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures S1, S2, S3, Table 1 and Table 2 (PDF 374 kb)
Rights and permissions
About this article
Cite this article
Angers, S., Thorpe, C., Biechele, T. et al. The KLHL12–Cullin-3 ubiquitin ligase negatively regulates the Wnt–β-catenin pathway by targeting Dishevelled for degradation. Nat Cell Biol 8, 348–357 (2006). https://doi.org/10.1038/ncb1381
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb1381
This article is cited by
-
New insights in ubiquitin-dependent Wnt receptor regulation in tumorigenesis
In Vitro Cellular & Developmental Biology - Animal (2024)
-
CYP2E1 plays a suppressive role in hepatocellular carcinoma by regulating Wnt/Dvl2/β-catenin signaling
Journal of Translational Medicine (2022)
-
Phosphorylation-induced changes in the PDZ domain of Dishevelled 3
Scientific Reports (2021)
-
RNA-Seq and differential gene expression analysis in Temora stylifera copepod females with contrasting non-feeding nauplii survival rates: an environmental transcriptomics study
BMC Genomics (2020)
-
Downregulation of the ubiquitin ligase KBTBD8 prevented epithelial ovarian cancer progression
Molecular Medicine (2020)