Abstract
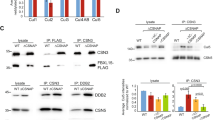
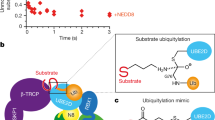
The COP9 signalosome (CSN) is known to bind cullin-RING ubiquitin ligases (CRLs) and to promote their activity in vivo1,2,3. The mechanism of this stimulation has remained enigmatic because CSN's intrinsic and associated enzymatic activities paradoxically inhibit CRL activity in vitro4,5. Reconciling this paradox, we show here that Csn5-catalysed cullin (Cul) deneddylation and Ubp12-mediated deubiquitination cooperate in maintaining the stability of labile substrate adapters, thus facilitating CRL function. Various fission-yeast csn and ubp12 deletion mutants have lower levels of the Cul3p adapter Btb3p. This decrease is due to increased autocatalytic, Cul3p–dependent, ubiquitination and the subsequent degradation of Btb3p. The CSN–Ubp12p pathway also maintains the stability of the Cul1p adapter Pop1p, a mechanism required for the efficient destruction of its cognate substrate Rum1p. Emphasizing the physiological importance of this mechanism, we found that the dispensable csn5 and ubp12 genes become essential for viability when adapter recruitment to Cul1p is compromised. Our data suggest that maintenance of adapter stability is a general mechanism of CRL control by the CSN.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Cope, G. A. & Deshaies, R. J. COP9 signalosome: a multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell 114, 663–671 (2003).
Wolf, D. A., Zhou, C. & Wee, S. The COP9 signalosome: an assembly and maintenance platform for cullin ubiquitin ligases? Nature Cell Biol. 5, 1029–1033 (2003).
Wei, N. & Deng, X. W. The COP9 signalosome. Annu. Rev. Cell Dev. Biol. 19, 261–286 (2003).
Groisman, R. et al. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 113, 357–367 (2003).
Zhou, C. et al. Fission yeast COP9/signalosome suppresses cullin activity through recruitment of the deubiquitylating enzyme Ubp12p. Mol. Cell 11, 927–938 (2003).
Deshaies, R. J. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15, 435–467 (1999).
Pintard, L. et al. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature 425, 311–316 (2003).
Xu, L. et al. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature 425, 316–321.
Geyer, R., Wee, S., Anderson, S., Yates, J. R. I. & Wolf, D. A. BTB/POZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol. Cell 12, 783–790 (2003).
Galan, J. M. & Peter, M. Ubiquitin-dependent degradation of multiple F-box proteins by an autocatalytic mechanism. Proc. Natl Acad. Sci. USA 96, 9124–9129 (1999).
Wirbelauer, C. et al. The F-box protein Skp2 is a ubiquitylation target of a Cul1-based core ubiquitin ligase complex: evidence for a role of Cul1 in the suppression of Skp2 expression in quiescent fibroblasts. EMBO J. 19, 5362–5375 (2000).
Zhou, P. & Howley, P. M. Ubiquitination and degradation of the substrate recognition subunits of SCF ubiquitin-protein ligases. Mol. Cell 2, 571–580 (1998).
Rouillon, A., Barbey, R., Patton, E. E., Tyers, M. & Thomas, D. Feedback-regulated degradation of the transcriptional activator Met4 is triggered by the SCF(Met30) complex. EMBO J. 19, 282–294 (2000).
Pan, Z. Q., Kentsis, A., Dias, D. C., Yamoah, K. & Wu, K. Nedd8 on cullin: building an expressway to protein destruction. Oncogene 23, 1985–1997 (2004).
Lyapina, S. et al. Promotion of NEDD8-CUL1 conjugate cleavage by COP9 signalosome. Science 292, 1382–1385 (2001).
Schwechheimer, C. et al. Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIRI in mediating auxin response. Science 292, 1379–1382 (2001).
Zhou, C. et al. The fission yeast COP9/signalosome is involved in cullin modification by ubiquitin-related Ned8p. BMC Biochemistry 2, 7 (2001).
Cope, G. et al. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of NEDD8 from CUL1. Science 298, 608–611 (2002).
Mundt, K. E., Liu, C. & Carr, A. M. Deletion mutants in COP9/signalosome subunits in fission yeast Schizosaccharomyces pombe display distinct phenotypes. Mol. Biol. Cell 13, 493–502 (2002).
Pintard, L. et al. Neddylation and deneddylation of CUL-3 is required to target MEI-1/katanin for degradation at the meiosis-to-mitosis transition in C. elegans. Curr. Biol. 13, 911–921 (2003).
Mundt, K. E. et al. The COP9/signalosome complex is conserved in fission yeast and has a role in S phase. Curr. Biol. 9, 1427–1430 (1999).
Kominami, K., Ochotorena, I. & Toda, T. Two F-box/WD-repeat proteins Pop1 and Pop2 form hetero- and homo- complexes together with cullin-1 in the fission yeast SCF (Skp1-Cullin-1-F-box) ubiquitin ligase. Genes Cells 3, 721–735 (1998).
Lehmann, A. et al. Molecular interactions of fission yeast Skp1 and its role in the DNA damage checkpoint. Genes Cells 9, 367–382 (2004).
Lu, L., Schulz, H. & Wolf, D. A. The F-box protein SKP2 mediates androgen control of p27 stability in LNCaP human prostate cancer cells. BMC Cell Biol. 3, 22 (2002).
Welcker, M. et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc. Natl Acad. Sci. USA 101, 9085–9090 (2004).
Deffenbaugh, A. E. et al. Release of ubiquitin charged Csc34-A-Ub from the RING domain is essential for ubiquitination of the SCFCdc4-bound substrate Sic1. Cell 114, 611–622 (2003).
Bahler, J. et al. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943–951 (1998).
Acknowledgements
We wish to thank S. Moreno for Rum1p antisera; G. Cope and R. Deshaies for csn5 JAMM mutants; and C. Zhou for assistance during much of these studies. This work was supported by NIH grant GM59780 to D.A.W., by grants from Cancer Research UK and the Human Frontier Science Program to T.T., and by the NIEHS Center Grant ES00002 and the NIEHS Training Grant ES07155.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary figures 1 and 2; supplementary table 1 (PDF 128 kb)
Rights and permissions
About this article
Cite this article
Wee, S., Geyer, R., Toda, T. et al. CSN facilitates Cullin–RING ubiquitin ligase function by counteracting autocatalytic adapter instability. Nat Cell Biol 7, 387–391 (2005). https://doi.org/10.1038/ncb1241
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb1241
This article is cited by
-
Regulatory mechanisms and therapeutic potential of JAB1 in neurological development and disorders
Molecular Medicine (2023)
-
Post-translational modifications: emerging regulators manipulating jasmonate biosynthesis and signaling
Plant Cell Reports (2022)
-
SlBBX20 interacts with the COP9 signalosome subunit SlCSN5-2 to regulate anthocyanin biosynthesis by activating SlDFR expression in tomato
Horticulture Research (2021)
-
CSN5 inhibition triggers inflammatory signaling and Rho/ROCK-dependent loss of endothelial integrity
Scientific Reports (2019)
-
COP9 signalosome subunit 5A affects phenylpropanoid metabolism, trichome formation and transcription of key genes of a regulatory tri-protein complex in Arabidopsis
BMC Plant Biology (2018)