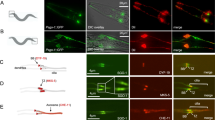
Abstract
The mechanisms that ensure centrosome duplication are poorly understood. In Caenorhabditis elegans, ZYG-1, SAS-4, SAS-5 and SPD-2 are required for centriole formation. However, it is unclear whether these proteins have functional homologues in other organisms. Here, we identify SAS-6 as a component that is required for daughter centriole formation in C. elegans. SAS-6 is a coiled-coil protein that is recruited to centrioles at the onset of the centrosome duplication cycle. Our analysis indicates that SAS-6 and SAS-5 associate and that this interaction, as well as ZYG-1 function, is required for SAS-6 centriolar recruitment. SAS-6 is the founding member of an evolutionarily conserved protein family that contains the novel PISA motif. We investigated the function of the human homologue of SAS-6. GFP–HsSAS-6 localizes to centrosomes and its overexpression results in excess foci-bearing centriolar markers. Furthermore, siRNA-mediated inactivation of HsSAS-6 in U2OS cells abrogates centrosome overduplication following aphidicolin treatment and interferes with the normal centrosome duplication cycle. Therefore, HsSAS-6 is also required for centrosome duplication, indicating that the function of SAS-6-related proteins has been widely conserved during evolution.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
O'Connell, K. F. et al. The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell 105, 547–558 (2001).
Kirkham, M., Müller-Reichert, T., Oegema, K., Grill, S. & Hyman, A. A. SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell 112, 575–587 (2003).
Leidel, S. & Gönczy, P. SAS-4 is essential for centrosome duplication in C. elegans and is recruited to daughter centrioles once per cell cycle. Dev. Cell 4, 431–439 (2003).
Delattre, M. et al. Centriolar SAS-5 is required for centrosome duplication in C. elegans. Nature Cell Biol. 6, 656–664 (2004).
Kemp, C. A., Kopish, K. R., Zipperlen, P., Ahringer, J. & O'Connell, K. F. Centrosome maturation and duplication in C. elegans require the coiled-coil protein SPD-2. Dev. Cell 6, 511–523 (2004).
Pelletier, L. et al. The Caenorhabditis elegans centrosomal protein SPD-2 is required for both pericentriolar material recruitment and centriole duplication. Curr. Biol. 14, 863–873 (2004).
Azimsadeh, J. & Bornens, M. in Centrosomes in Development and Disease (ed. Nigg, E. A.) 93–122 (Wiley-VCH, Weinheim, 2004).
Wolf, N., Hirsh, D. & McIntosh, J. R. Spermatogenesis in males of the free-living nematode, Caenorhabditis elegans. J. Ultrastruct. Res. 63, 155–169 (1978).
Salisbury, J. L., Suino, K. M., Busby, R. & Springett, M. Centrin-2 is required for centriole duplication in mammalian cells. Curr. Biol. 12, 1287–1292 (2002).
Dutcher, S. K. & Trabuco, E. C. The UNI3 gene is required for assembly of basal bodies of Chlamydomonas and encodes δ-tubulin, a new member of the tubulin superfamily. Mol. Biol. Cell 9, 1293–1308 (1998).
Dutcher, S. K., Morrissette, N. S., Preble, A. M., Rackley, C. & Stanga, J. ε-tubulin is an essential component of the centriole. Mol. Biol. Cell 13, 3859–3869 (2002).
Garreau de Loubresse, N., Ruiz, F., Beisson, J. & Klotz, C. Role of δ-tubulin and the C-tubule in assembly of Paramecium basal bodies. BMC Cell Biol. 2, 4 Epub 2001 Mar 07 (2001).
Hung, L. Y., Tang, C. J. & Tang, T. K. Protein 4.1 R-135 interacts with a novel centrosomal protein (CPAP) which is associated with the γ-tubulin complex. Mol Cell Biol. 20, 7813–7825 (2000).
Andersen, J. S. et al. Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426, 570–574 (2003).
Piano, F., Schetter, A. J., Mangone, M., Stein, L. & Kemphues, K. J. RNAi analysis of genes expressed in the ovary of Caenorhabditis elegans. Curr. Biol. 10, 1619–1622 (2000).
Sönnichsen, B. et al. Full genome RNAi profiling of early embryogenesis in C. elegans. Nature (in the press).
Bellanger, J. M. & Gönczy, P. TAC-1 and ZYG-9 form a complex that promotes microtubule assembly in C. elegans embryos. Curr. Biol. 13, 1488–1498 (2003).
Wolff, A. et al. Distribution of glutamylated α and β-tubulin in mouse tissues using a specific monoclonal antibody, GT335. Eur. J. Cell Biol. 59, 425–432 (1992).
Meraldi, P., Honda, R. & Nigg, E. A. Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53−/− cells. EMBO J. 21, 483–492 (2002).
Meraldi, P., Lukas, J., Fry, A. M., Bartek, J. & Nigg, E. A. Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-cyclin A. Nature Cell Biol. 1, 88–93 (1999).
Warnke, S. et al. Polo-like kinase-2 is required for centriole duplication in mammalian cells. Curr. Biol. 14, 1200–1207 (2004).
Chase, D. et al. The polo-like kinase PLK-1 is required for nuclear envelope breakdown and the completion of meiosis in Caenorhabditis elegans. Genesis 26, 26–41 (2000).
Pihan, G. A., Wallace, J., Zhou, Y. & Doxsey, S. J. Centrosome abnormalities and chromosome instability occur together in pre-invasive carcinomas. Cancer Res. 63, 1398–1404 (2003).
Dammermann, A., Mü ller-Reichert, T., Pelletier, L., Habermann, B., Desai, A. & Oegema, K. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev. Cell 7, 815–829 (2004).
Brenner, S. The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974).
Wood, W. B. et al. Parental effects and phenotypic characterization of mutations that affect early development in Caenorhabditis elegans. Dev. Biol. 74, 446–469 (1980).
Nelson, G. A., Lew, K. K. & Ward, S. Intersex, a temperature-sensitive mutant of the nematode Caenorhabditis elegans. Dev. Biol. 66, 386–409 (1978).
Timmons, L., Court, D. L. & Fire, A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263, 103–112 (2001).
Praitis, V., Casey, E., Collar, D. & Austin, J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157, 1217–1226 (2001).
Pagni, M. et al. MyHits: a new interactive resource for protein annotation and domain identification. Nucleic Acids Res. 32, W332–W335 (2004).
Sigrist, C. J. et al. PROSITE: a documented database using patterns and profiles as motif descriptors. Brief Bioinform. 3, 265–274 (2002).
Braun, P. et al. Proteome-scale purification of human proteins from bacteria. Proc. Natl Acad. Sci. USA 99, 2654–2659 (2002).
Fry, A. M. et al. C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J. Cell Biol. 141, 1563–1574 (1998).
Middendorp, S. et al. A role for centrin 3 in centrosome reproduction. J. Cell Biol. 148, 405–416 (2000).
Pintard, L. et al. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature 425, 311–316 (2003).
Clamp, M., Cuff, J., Searle, S. M. & Barton, G. J. The Jalview Java alignment editor. Bioinformatics 20, 426–427 Epub 2004 Jan 2022 (2004).
Acknowledgements
We are especially grateful to X. Yan, R. Habedanck and E. Nigg for guidance with cell culture techniques and reagents, as well as M. Cockell for help in generating SAS-6 fusion proteins. We thank J. Azimsadeh, M. Bornens, B. Eddé, M. Glotzer and V. Simanis for reagents. C. Echeverri is acknowledged for communicating results before publication; R. Iggo, V. Simanis and P. Strnad for critical reading of the manuscript; M. Migliaccio, P. Strnad and A. Wilson for help with FACS analysis; C. Bonnard and N. Garin for microscopy support; and D. Moersch and I. Jouravleff for helpful discussions. Some strains were obtained from the Caenorhabditis Genetics Center, which is funded by the National Institute of Health National Center for Research Resources (NCRR). M. D. is recipient of a Roche Research Foundation postdoctoral fellowship. Oncosuisse supports work on centrosome duplication in the Gönczy laboratory (OCS-01495-02-2004).
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary information
Fig S1, legends accompanying movies (PDF 140 kb)
Rights and permissions
About this article
Cite this article
Leidel, S., Delattre, M., Cerutti, L. et al. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat Cell Biol 7, 115–125 (2005). https://doi.org/10.1038/ncb1220
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb1220
This article is cited by
-
A novel HIF1α-STIL-FOXM1 axis regulates tumor metastasis
Journal of Biomedical Science (2022)
-
Tuning SAS-6 architecture with monobodies impairs distinct steps of centriole assembly
Nature Communications (2021)
-
Primary microcephaly with an unstable genome
Genome Instability & Disease (2020)
-
A dynamically interacting flexible loop assists oligomerisation of the Caenorhabditis elegans centriolar protein SAS-6
Scientific Reports (2019)
-
Caspase-mediated cleavage of the centrosomal proteins during apoptosis
Cell Death & Disease (2018)