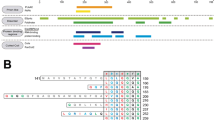
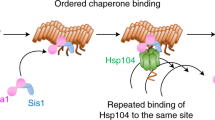
Abstract
Mammalian and most fungal infectious proteins (also known as prions) are self-propagating amyloid, a filamentous β-sheet structure. A prion domain determines the infectious properties of a protein by forming the core of the amyloid. We compare the properties of known prion domains and their interactions with the remainder of the protein and with chaperones. Ure2p and Sup35p, two yeast prion proteins, can still form prions when the prion domains are shuffled, indicating a parallel in-register β-sheet structure.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Griffith, J. S. Self-replication and scrapie. Nature 215, 1043–1044 (1967).
Prusiner, S. B. Novel proteinaceous infectious particles cause scrapie. Science 216, 136–144 (1982).
Tuite, M. F. & Koloteva-Levin, N. Propagating prions in fungi and mammals. Mol. Cell 14, 541–552 (2004).
Chernoff, Y. O. Amyloidogenic domains, prions and structural inheritance: rudiments of early life or recent acquisition? Curr. Opin. Chem. Biol. 8, 665–671 (2004).
Wickner, R. B. et al. Prions: proteins as genes and infectious entities. Genes Dev. 18, 470–485 (2004).
Aguzzi, A. & Polymenidou, M. Mammalian prion biology: one century of evolving concepts. Cell 116, 313–327 (2004).
Prusiner, S. B. Prions. Proc. Natl Acad. Sci. USA 95, 13363–13383 (1998).
Wickner, R. B. [URE3] as an altered URE2 protein: evidence for a prion analog in S. cerevisiae. Science 264, 566–569 (1994).
Derkatch, I. L., Chernoff, Y. O., Kushnirov, V. V., Inge-Vechtomov, S. G. & Liebman, S. W. Genesis and variability of [PSI ] prion factors in Saccharomyces cerevisiae. Genetics 144, 1375–1386 (1996).
Coustou, V., Deleu, C., Saupe, S. & Begueret, J. The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc. Natl Acad. Sci. USA 94, 9773–9778 (1997).
Caughey, B. & Lansbury, P. T. Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible prion aggregates from innocent bystanders. Annu. Rev. Neurosci. 26, 267–298 (2003).
Magasanik, B. & Kaiser, C. A. Nitrogen regulation in Saccharomyces cerevisiae. Gene 290, 1–18 (2002).
Liebman, S. W. & Derkatch, I. L. The yeast [PSI+] prion: making sense out of nonsense. J. Biol. Chem. 274, 1181–1184 (1999).
Derkatch, I. L., Bradley, M. E., Hong, J. Y. & Liebman, S. W. Prions affect the appearance of other prions: the story of [PIN]. Cell 106, 171–182 (2001).
Saupe, S. J. Molecular genetics of heterokaryon incompatibility in filamentous ascomycetes. Microbiol. Mol. Biol. Rev. 64, 489–502 (2000).
Roberts, B. T. & Wickner, R. B. A class of prions that propagate via covalent auto-activation. Genes Dev. 17, 2083–2087 (2003).
Kicka, S. & Silar, P. PaASK1, a mitogen-activated protein kinase kinase kinase that controls cell degeneration and cell differentiation in Podospora anserina. Genetics 166, 1241–1252 (2004).
Masison, D. C. & Wickner, R. B. Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science 270, 93–95 (1995).
Taylor, K. L., Cheng, N., Williams, R. W., Steven, A. C. & Wickner, R. B. Prion domain initiation of amyloid formation in vitro from native Ure2p. Science 283, 1339–1343 (1999).
TerAvanesyan, A., Dagkesamanskaya, A. R., Kushnirov, V. V. & Smirnov, V. N. The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [psi+] in the yeast Saccharomyces cerevisiae. Genetics 137, 671–676 (1994).
King, C. -Y. et al. Prion-inducing domain 2–114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc. Natl Acad. Sci. USA 94, 6618–6622 (1997).
Glover, J. R. et al. Self-seeded fibers formed by Sup35, the protein determinant of [PSI +], a heritable prion-like factor of S. cerevisiae. Cell 89, 811–819 (1997).
Bolton, D. C., McKinley, M. P. & Prusiner, S. B. Identification of a protein that purifies with the scrapie prion. Science 218, 1309–1311 (1982).
Fischer, M. et al. Prion protein (PrP) with amino-terminal deletions restoring susceptibility of PrP knockout mice to scrapie. EMBO J. 15, 1255–1264 (1996).
Balguerie, A. et al. Domain organization and structure-function relationship of the HET-s prion protein of Podospora anserina. EMBO J. 22, 2071–2081 (2003).
Baxa, U. et al. Architecture of Ure2p prion filaments: the N-terminal domain forms a central core fiber. J. Biol. Chem. 278, 43717–43727 (2003).
Maddelein, M. L., Dos Reis, S., Duvezin-Caubet, S., Coulary-Salin, B. & Saupe, S. J. Amyloid aggregates of the HET-s prion protein are infectious. Proc. Natl Acad. Sci. USA 99, 7402–7407 (2002).
King, C. Y. & Diaz-Avalos, R. Protein-only transmission of three yeast prion strains. Nature 428, 319–323 (2004).
Tanaka, M., Chien, P., Naber, N., Cooke, R. & Weissman, J. S. Conformational variations in an infectious protein determine prion strain differences. Nature 428, 323–328 (2004).
Brachmann, A., Baxa, U. & Wickner, R. B. Prion generation in vitro: amyloid of Ure2p is infectious. EMBO J. 24, 3082–3092 (2005).
Legname, G. et al. Synthetic mammalian prions. Science 305, 673–676 (2004).
Doel, S. M., McCready, S. J., Nierras, C. R. & Cox, B. S. The dominant PNM2− mutation which eliminates the [PSI] factor of Saccharomyces cerevisiae is the result of a missense mutation in the SUP35 gene. Genetics 137, 659–670 (1994).
DePace, A. H., Santoso, A., Hillner, P. & Weissman, J. S. A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell 93, 1241–1252 (1998).
Kochneva-Pervukhova, N. V. et al. Mechanism of inhibition of Ψ+ prion determinant propagation by a mutation of the N-terminus of the yeast Sup35 protein. EMBO J. 17, 5805–5810 (1998).
Derkatch, I. L., Bradley, M. E., Zhou, P. & Liebman, S. W. The PNM2 mutation in the prion protein domain of SUP35 has distinct effects on different variants of the [PSI +] prion in yeast. Curr. Genet. 35, 59–67 (1999).
Mead, S. et al. Balancing selection at the prion protein gene consistent with prehistoric kurulike epidemics. Science 300, 640–643 (2003).
Scott, M. et al. Propagation of prions with artificial properties in transgenic mice expressing chimeric PrP genes. Cell 73, 979–988 (1993).
Priola, S. A., Caughey, B., Race, R. E. & Chesebro, B. Heterologous PrP molecules interfere with accumulation of protease-resistant PrP in scrapie-infected murine neuroblastoma cells. J. Virol. 68, 4873–4878 (1994).
Prusiner, S. B. et al. Transgenic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell 63, 673–686 (1990).
Edskes, H. K., Gray, V. T. & Wickner, R. B. The [URE3] prion is an aggregated form of Ure2p that can be cured by overexpression of Ure2p fragments. Proc. Natl Acad. Sci. USA 96, 1498–1503 (1999).
Owen, F. et al. Insertion in prion protein gene in familial Creutzfeldt–Jakob disease. Lancet 1, 51–52 (1989).
Liu, J. J. & Lindquist, S. Oligopeptide-repeat expansions modulate 'protein-only' inheritance in yeast. Nature 400, 573–576 (1999).
Osherovich, L. Z., Cox, B. S., Tuite, M. F. & Weissman, J. S. Dissection and design of yeast prions. PLoS Biol 2, E86 (2004).
Flechsig, E. et al. Prion protein devoid of the octapeptide repeat region restores susceptibility to scrapie in PrP knockout mice. Neuron 27, 399–408 (2000).
Prusiner, S. B., Groth, D. F., Bolton, D. C., Kent, S. B. & Hood, L. E. Purification and structural studies of a major scrapie prion protein. Cell 38, 127–134 (1984).
Parham, S. N., Resende, C. G. & Tuite, M. F. Oligopeptide repeats in the yeast protein Sup35p stabilize intermolecular prion interactions. EMBO J. 20, 2111–2119 (2001).
Borchsenius, A. S., Wegrzyn, R. D., Newnam, G. P., Inge-Vechtomov, S. G. & Chernoff, Y. O. Yeast prion protein derivative defective in aggregate shearing and production of new 'seeds'. EMBO J. 20, 6683–6691 (2001).
Maddelein, M. -L. & Wickner, R. B. Two prion-inducing regions of Ure2p are non-overlapping. Mol. Cell. Biol. 19, 4516–4524 (1999).
Fernandez-Bellot, E., Guillemet, E. & Cullin, C. The yeast prion [URE3] can be greatly induced by a functional mutated URE2 allele. EMBO J. 19, 3215–3222 (2000).
Ross, E. D., Baxa, U. & Wickner, R. B. Scrambled prion domains form prions and amyloid. Mol. Cell. Biol. 24, 7206–7213 (2004).
Ross, E. D., Edskes, H. K., Terry, M. J. & Wickner, R. B. Primary sequence independence for prion formation. Proc. Natl Acad. Sci. USA 102, 12825–12830 (2005).
Antzutkin, O. N., Leapman, R. D., Balbach, J. J. & Tycko, R. Supramolecular structural constraints on Alzheimer's β-amyloid fibrils from electron microscopy and solid-state nuclear magnetic resonance. Biochemistry 41, 15436–15450 (2002).
Perutz, M. F., Johnson, T., Suzuki, M. & Finch, J. T. Glutamine repeats as polar zippers: their possible role in inherited neurodegenerative diseases. Proc. Natl Acad. Sci. USA 91, 5355–5358 (1994).
Bradley, M. E., Edskes, H. K., Hong, J. Y., Wickner, R. B. & Liebman, S. W. Interactions among prions and prion “strains” in yeast. Proc. Natl Acad. Sci. USA 99 (Suppl. 4), 16392–16399 (2002).
Derkatch, I. L. et al. Effects of Q/N-rich, polyQ, and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro. Proc. Natl Acad. Sci. USA 101, 12934–12939 (2004).
Salnikova, A. B., Kryndushkin, D. S., Smirnov, V. N., Kushnirov, V. V. & Ter-Avanesyan, M. D. Nonsense suppression in yeast cells overproducing Sup35 (eRF3) is caused by its non-heritable amyloids. J. Biol. Chem. 280, 8808–8812 (2005).
Benzinger, T. L. et al. Propagating structure of Alzheimer's β-amyloid(10–35) is parallel β-sheet with residues in exact register. Proc. Natl Acad. Sci. USA 95, 13407–13412 (1998).
Antzutkin, O. N. et al. Multiple quantum solid-state NMR indicates a parallel, not antiparallel, organization of β-sheets in Alzheimer's β-amyloid fibrils. Proc. Natl Acad. Sci. USA 97, 13045–13050 (2000).
Tycko, R. Insights into the amyloid folding problem from solid-state NMR. Biochemistry 42, 3151–3159 (2003).
Chan, J. C. C., Oyler, N. A., Yau, W. -M. & Tycko, R. Parallel β-sheets and polar zippers in amyloid fibrils formed by residues 10–39 of the yeast prion protein Ure2p. Biochemistry 44, 10669–10680 (2005).
Jayasinghe, S. A. & Langen, R. Identifying structural features of fibrillar islet amyloid polypeptide using site-directed spin labeling. J. Biol. Chem. 279, 48420–48425 (2004).
Der-Sarkissian, A., Jao, C. C., Chen, J. & Langen, R. Structural organization of α-synuclein fibrils studied by site-directed spin labeling. J. Biol. Chem. 278, 37530–37535 (2003).
Balbach, J. J. et al. Amyloid fibril formation by Aβ16–22, a seven-residue fragment of the Alzheimer's β-amyloid peptide, and structural characterization by solid state NMR. Biochemistry 39, 13748–13759 (2000).
Govaerts, C., Wille, H., Prusiner, S. B. & Cohen, F. E. Evidence for assembly of prions with left-handed β-helices into trimers. Proc. Natl Acad. Sci. USA 101, 8342–8347 (2004).
Serio, T. R. et al. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science 289, 1317–1321 (2000).
Kishimoto, A. et al. β-Helix is a likely core structure of yeast prion Sup35 amyloid fibers. Biochem. Biophys. Res. Commun. 315, 739–745 (2004).
Nelson, R. et al. Structure of the cross-β spine of amyloid-like fibrils. Nature 435, 773–778 (2005).
Krishnan, R. & Lindquist, S. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature 435, 765–772 (2005).
DePace, A. H. & Weissman, J. S. Origins and kinetic consequences of diversity in Sup35 yeast prion fibers. Nature Struct. Biol. 9, 389–396 (2002).
Inoue, Y., Kishimoto, A., Hirao, J., Yoshida, M. & Taguchi, H. Strong growth polarity of yeast prion fiber revealed by single fiber imaging. J. Biol. Chem. 276, 35227–35230 (2001).
Pierce, M. M., Baxa, U., Steven, A. C., Bax, A. & Wickner, R. B. Is the prion domain of soluble Ure2p unstructured? Biochemistry 44, 321–328 (2005).
Baxa, U., Speransky, V., Steven, A. C. & Wickner, R. B. Mechanism of inactivation on prion conversion of the Saccharomyces cerevisiae Ure2 protein. Proc. Natl Acad. Sci. USA 99, 5253–5260 (2002).
Bousset, L., Belrhali, H., Melki, R. & Morera, S. Crystal structures of the yeast prion Ure2p functional region in complex with glutathione and related compounds. Biochemistry 40, 13564–13573 (2001).
Bai, M., Zhou, J. M. & Perrett, S. The yeast prion protein Ure2 shows glutathione peroxidase activity in both native and fibrillar forms. J. Biol. Chem. 279, 50025–50030 (2004).
Schlumpberger, M. et al. The prion domain of yeast Ure2p induces autocatalytic formation of amyloid fibers by a recombinant fusion protein. Protein Sci. 9, 440–451 (2000).
Baxa, U. et al. Filaments of the Ure2p prion protein have a cross-β core structure. J. Struct. Biol. 150, 170–179 (2005).
Kochneva-Pervukhova, N. V., Poznyakovski, A. I., Smirnov, V. N. & Ter-Avanesyan, M. D. C-terminal truncation of the Sup35 protein increases the frequency of de novo generation of a prion-based [PSI+ ] determinant in Saccharmyces cerevisiae. Curr. Genet. 34, 146–151 (1998).
Brown, P. et al. Human spongiform encephalopathy: the National Institutes of Health series of 300 cases of experimentally transmitted disease. Ann. Neurol. 35, 513–529 (1994).
Locht, C., Chesebro, B., Race, R. & Keith, J. M. Molecular cloning and complete sequence of prion protein cDNA from mouse brain infected with the scrapie agent. Proc. Natl Acad. Sci. USA 83, 6372–6376 (1986).
Basler, K. et al. Scrapie and cellular PrP isoforms are encoded by the same chromosomal locus. Cell 46, 417–428 (1986).
Riek, R., Hornemann, S., Wider, G., Glockshuber, R. & Wuthrich, K. NMR characterization of the full-length recombinant murine prion protein, mPrP(23–231). FEBS Lett. 413, 282–288 (1997).
Chernoff, Y. O., Lindquist, S. L., Ono, B. -I., Inge-Vechtomov, S. G. & Liebman, S. W. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 268, 880–884 (1995).
Derkatch, I. L., Bradley, M. E., Zhou, P., Chernoff, Y. O. & Liebman, S. W. Genetic and environmental factors affecting the de novo appearance of the [PSI +] prion in Saccharomyces cerevisiae. Genetics 147, 507–519 (1997).
Moriyama, H., Edskes, H. K. & Wickner, R. B. [URE3] prion propagation in Saccharomyces cerevisiae: requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol. Cell. Biol. 20, 8916–8922 (2000).
Ness, F., Ferreira, P., Cox, B. S. & Tuite, M. F. Guanidine hydrochloride inhibits the generation of prion “seeds” but not prion protein aggregation in yeast. Mol. Cell. Biol. 22, 5593–5605 (2002).
Jung, G., Jones, G. & Masison, D. C. Amino acid residue 184 of yeast Hsp104 chaperone is critical for prion-curing by guanidine, prion propagation, and thermotolerance. Proc. Natl Acad. Sci. USA 99, 9936–9941 (2002).
Paushkin, S. V., Kushnirov, V. V., Smirnov, V. N. & Ter-Avanesyan, M. D. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 15, 3127–3134 (1996).
Wegrzyn, R. D., Bapat, K., Newnam, G. P., Zink, A. D. & Chernoff, Y. O. Mechanism of prion loss after Hsp104 inactivation in yeast. Mol. Cell. Biol. 21, 4656–4669 (2001).
Glover, J. R. & Lindquist, S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94, 73–82 (1998).
Newnam, G. P., Wegrzyn, R. D., Lindquist, S. L. & Chernoff, Y. O. Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol. Cell. Biol. 19, 1325–1333 (1999).
Kushnirov, V. V., Kryndushkin, D. S., Boguta, M., Smirnov, V. N. & Ter-Avanesyan, M. D. Chaperones that cure yeast artificial [PSI+] and their prion-specific effects. Curr. Biol. 10, 1443–1446 (2000).
Jung, G., Jones, G., Wegrzyn, R. D. & Masison, D. C. A role for cytosolic Hsp70 in yeast [PSI+] prion propagation and [PSI+] as a cellular stress. Genetics 156, 559–570 (2000).
Allen, K. D. et al. Hsp70 chaperones as modulators of prion life cycle: novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+]. Genetics 169, 1227–1242 (2005).
Shorter, J. & Lindquist, S. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science 304, 1793–1797 (2004).
Inoue, Y., Taguchi, H., Kishimoto, A. & Yoshida, M. Hsp104 binds to yeast Sup35 prion fiber but needs other factor(s) to sever it. J. Biol. Chem. 279, 52319–52323 (2004).
Crist, C. G., Nakayashiki, T., Kurahashi, H. & Nakamura, Y. [PHI+], a novel Sup35-prion variant propagated with non-Gln/Asn oligopeptide repeats in the absence of the chaperone protein Hsp104. Genes Cells 8, 603–618 (2003).
Bruce, M. E., McConnell, I., Fraser, H. & Dickinson, A. G. The disease characteristics of different strains of scrapie in Sinc congenic mouse lines: implications for the nature of the agent and host control of pathogenesis. J. Gen. Virol. 72, 595–603 (1991).
Chien, P., Weissman, J. S. & DePace, A. H. Emerging principles of conformation-based prion inheritance. Annu. Rev. Biochem. 73, 617–656 (2004).
Collinge, J. Variant Creutzfeldt–Jakob disease. Lancet 354, 317–323 (1999).
Hatters, D. M., Minton, A. P. & Howlett, G. J. Macromolecular crowding accelerates amyloid formation by human apolipoprotein C-II. J. Biol. Chem. 277, 7824–7830 (2002).
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Ross, E., Minton, A. & Wickner, R. Prion domains: sequences, structures and interactions. Nat Cell Biol 7, 1039–1044 (2005). https://doi.org/10.1038/ncb1105-1039
Issue Date:
DOI: https://doi.org/10.1038/ncb1105-1039
This article is cited by
-
Discovering putative prion sequences in complete proteomes using probabilistic representations of Q/N-rich domains
BMC Genomics (2013)
-
Drosophila GAGA factor polyglutamine domains exhibit prion-like behavior
BMC Genomics (2013)
-
Foreword to the biophysics of protein-protein and protein-ligand interactions in dilute and crowded media—a special issue in honor of Allen Minton’s 70th birthday
Biophysical Reviews (2013)
-
Computational modeling of the relationship between amyloid and disease
Biophysical Reviews (2012)