Abstract
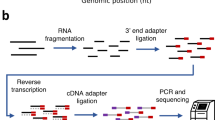
Identification of the mRNA start site is essential in establishing the full-length cDNA sequence of a gene and analyzing its promoter region, which regulates gene expression. Here we describe the development of a 5′-end serial analysis of gene expression (5′ SAGE) that can be used to globally identify transcriptional start sites and the frequency of individual mRNAs. Of the 25,684 5′ SAGE tags in the HEK293 human cell library, 19,893 matched to the human genome. Among 15,448 tags in one locus of the genome, 85.8%–96.1% of the 5′ SAGE tags were assigned within −500 to +200 nt of mRNA start sites using the RefSeq, UniGene and DBTSS databases. This technique should facilitate 5′-end transcriptome analysis in a variety of cells and tissues.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Duggan, D.J., Bittner, M., Chen, Y., Meltzer, P. & Trent, J.M. Expression profiling using cDNA microarrays. Nat. Genet. 21, 10–14 (1999).
Saha, S. et al. Using the transcriptome to annotate the genome. Nat. Biotechnol. 20, 508–512 (2002).
Madden, S.L., Galella, E.A., Zhu, J., Bertelsen, A.H. & Beaudry, G.A. SAGE transcript profiles for p53-dependent growth regulation. Oncogene 15, 1079–1085 (1997).
Velculescu, V.E. et al. Analysis of human transcriptomes. Nat. Genet. 23, 387–388 (1999).
Hashimoto, S. et al. Gene expression profile in human leukocytes. Blood 101, 3509–3513 (2003).
Lander, E.S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Maruyama, K. & Sugano, S. Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides. Gene 138, 171–174 (1994).
Suzuki, Y. et al. DBTSS: DataBase of human Transcriptional Start Sites and full-length cDNAs. Nucleic Acids Res. 30, 328–331 (2002).
Suzuki, Y. et al. Diverse transcriptional initiation revealed by fine, large-scale mapping of mRNA start sites. EMBO Rep. 2, 388–393 (2001).
Pauws, E., van Kampen, A.H., van de Graaf, S.A., de Vijlder, J.J. & Ris-Stalpers, C. Heterogeneity in polyadenylation cleavage sites in mammalian mRNA sequences: implications for SAGE analysis. Nucleic Acids Res. 29, 1690–1694 (2001).
Shiraki, T. et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc. Natl. Acad. Sci. USA 100, 15776–15781 (2003).
Modrek, B. & Lee, C. A genomic view of alternative splicing. Nat. Genet. 30, 13–19 (2002).
Krawczak, M., Reiss, J. & Cooper, D.N. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum. Genet. 90, 41–54 (1992).
Zavolan, M. et al. Impact of alternative initiation, splicing, and termination on the diversity of the mRNA transcripts encoded by the mouse transcriptome. Genome Res. 13, 1290–1300 (2003).
Hashimoto, S.-I., Suzuki, T., Dong, H.-Y., Yamazaki, N. & Matsushima, K. Serial analysis of gene expression in human monocytes and macrophages. Blood 94, 837–844 (1999).
Suzuki, Y., Yoshitomo-Nakagawa, K., Maruyama, K., Suyama, A. & Sugano, S. Construction and characterization of a full length-enriched and a 5′-end-enriched cDNA library. Gene 200, 149–156 (1997).
Honkura, T., Ogasawara, J., Yamada, T. & Morishita, S. The Gene Resource Locator: gene locus maps for transcriptome analysis. Nucleic Acids Res. 30, 221–225 (2002).
Wheeler, D.L. Database Resources of the National Center for Biotechnology. Nucleic Acids Res. 31, 28–33 (2003).
Acknowledgements
This work was supported by Grant-in-Aid for Scientific Research on Priority Areas (C) “Medical Genome Science” from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Comparison of the novel 5′ ends of representative known genes between 5′SAGE and the directly sequenced data of the 5′ end of captured full length cDNAs in HEK293. (PDF 12 kb)
Supplementary Fig. 2
Scatter plot of the frequency of 5′SAGE and 3′SAGE tags. (PDF 115 kb)
Supplementary Table 1
Identification of uncharacterized candidate genes and exons. (PDF 6 kb)
Supplementary Table 2
Profile of the 5′-end transcripts in HEK293 cells. (PDF 10 kb)
Rights and permissions
About this article
Cite this article
Hashimoto, Si., Suzuki, Y., Kasai, Y. et al. 5′-end SAGE for the analysis of transcriptional start sites. Nat Biotechnol 22, 1146–1149 (2004). https://doi.org/10.1038/nbt998
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt998
This article is cited by
-
Quantitative analysis of transcription start site selection reveals control by DNA sequence, RNA polymerase II activity and NTP levels
Nature Structural & Molecular Biology (2024)
-
Alternative transcription start sites of the enolase-encoding gene enoA are stringently used in glycolytic/gluconeogenic conditions in Aspergillus oryzae
Current Genetics (2020)
-
EXPRSS: an Illumina based high-throughput expression-profiling method to reveal transcriptional dynamics
BMC Genomics (2014)
-
5'-Serial Analysis of Gene Expression studies reveal a transcriptomic switch during fruiting body development in Coprinopsis cinerea
BMC Genomics (2013)
-
Transcriptome analysis of the filamentous fungus Aspergillus nidulans directed to the global identification of promoters
BMC Genomics (2013)