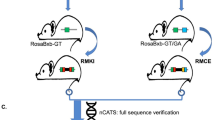
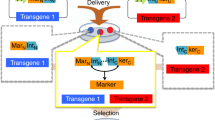
Abstract
The mouse is the leading vertebrate model because its genome can be altered by both random transgenesis and homologous recombination with targeting constructs. Both approaches have been hindered by the size and site limitations implicit in conventional Escherichia coli DNA-engineering methods. Homologous recombination in E. coli, or 'recombineering', has overcome these limitations for bacterial artificial chromosome (BAC) transgenesis1,2,3. Here we applied Red/ET recombineering (using the lambda Redα/Redβ recombinase pair)4,5,6 to generate a 64 kilobase targeting construct that carried two selectable cassettes permitting the simultaneous mutation of the target gene, Mll, at sites 43 kb apart in one round of mouse embryonic stem (ES) cell targeting. The targeting frequency after dual selection was 6%. Because the two selectable cassettes were flanked by FRT or loxP sites, three more alleles can be generated by site-specific recombination. Our approach represents a simple way to introduce changes at two or more sites in a genetic locus, and thereafter generate allele combinations. The size of BAC templates offers new freedom for the design of targeting constructs. Combined with the use of two selectable cassettes placed far apart, BAC-based targeting constructs may be applicable to tasks such as regional exchanges, deletions, and insertions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Muyrers, J.P., Zhang, Y. & Stewart, A.F. Techniques: recombinogenic engineering—new options for cloning and manipulating DNA. Trends Biochem. Sci. 26, 325–331 (2001).
Copeland, N.G., Jenkins, N.A. & Court, D.L. Recombineering: a powerful new tool for mouse functional genomics. Nat. Rev. Genet. 2, 769–779 (2001).
Heintz, N. BAC to the future: the use of bac transgenic mice for neuroscience research. Nat. Rev. Neurosci. 2, 861–870 (2001).
Zhang, Y., Buchholz, F., Muyrers, J.P. & Stewart, A.F. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 20, 123–128 (1998).
Muyrers, J.P., Zhang, Y., Testa, G. & Stewart, A.F. Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res. 27, 1555–1557 (1999).
Zhang, Y., Muyrers, J.P., Testa, G. & Stewart, A.F. DNA cloning by homologous recombination in Escherichia coli. Nat. Biotechnol. 18, 1314–1317 (2000).
Testa, G. & Stewart, A.F. Creating a translocation. Engineering interchromosomal translocations in the mouse. EMBO Rep. 1, 120–121 (2000).
Rowley, J.D. The critical role of chromosome translocations in human leukemias. Annu. Rev. Genet. 32, 495–519 (1998).
Nilson, I. et al. Exon/intron structure of the human ALL-1 (MLL) gene involved in translocations to chromosomal region 11q23 and acute leukaemias. Br. J. Haematol. 93, 966–972 (1996).
Angrand, P.O., Daigle, N., van der Hoeven, F., Scholer, H.R. & Stewart, A.F. Simplified generation of targeting constructs using ET recombination. Nucleic Acids Res. 27, e16 (1999).
Rigaut, G. et al. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030–1032 (1999).
Buchholz, F., Angrand, P.O. & Stewart, A.F. Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat. Biotechnol. 16, 657–662 (1998).
Zambrowicz, B.P. et al. Disruption of overlapping transcripts in the ROSA βgeo26 gene trap strain leads to widespread expression of β-galactosidase in mouse embryos and hematopoietic cells. Proc. Natl. Acad. Sci. USA 94, 3789–3794 (1997).
Moens, C.B., Auerbach, A.B., Conlon, R.A., Joyner, A.L. & Rossant, J. A targeted mutation reveals a role for N-myc in branching morphogenesis in the embryonic mouse lung. Genes Dev. 6, 691–704 (1992).
Muyrers, J.P. et al. Point mutation of bacterial artificial chromosomes by ET recombination. EMBO Rep. 1, 239–243 (2000).
Ellis, H.M., Yu, D., DiTizio, T. & Court, D.L. High-efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc. Natl. Acad. Sci. USA 98, 6742–6746 (2001).
Baer, A. & Bode, J. Coping with kinetic and thermodynamic barriers: RMCE, an efficient strategy for the targeted integration of transgenes. Curr. Opin. Biotechnol. 12, 473–480 (2001).
Meyers, E.N., Lewandoski, M. & Martin, G.R. An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nat. Genet. 18, 136–141 (1998).
Nagy, A. et al. Dissecting the role of N-myc in development using a single targeting vector to generate a series of alleles. Curr. Biol. 8, 661–664 (1998).
Andreas, S., Schwenk, F., Kuter-Luks, B., Faust, N. & Kuhn, R. Enhanced efficiency through nuclear localization signal fusion on phage PhiC31-integrase: activity comparison with Cre and FLPe recombinase in mammalian cells. Nucleic Acids Res. 30, 2299–2306 (2002).
Buchholz, F. & Stewart, A.F. Alteration of Cre recombinase site specificity by substrate-linked protein evolution. Nat. Biotechnol. 19, 1047–1052 (2001).
Ayton, P. et al. Truncation of the Mll gene in exon 5 by gene targeting leads to early preimplantation lethality of homozygous embryos. Genesis 30, 201–212 (2001).
Benes, V., Kilger, C., Voss, H., Paabo, S. & Ansorge, W. Direct primer walking on P1 plasmid DNA. Biotechniques 23, 98–100 (1997).
Nichols, J., Evans, E.P. & Smith, A.G. Establishment of germ-line-competent embryonic stem (ES) cells using differentiation-inhibiting activity. Development 110, 1341–1348 (1990).
Joyner, A.L. (ed.). Gene Targeting. A Practical Approach (Oxford University Press, New York, 2000).
Acknowledgements
We wish to thank Konstantinos Anastassiadis, William Brown, Frank van der Hoeven, Robin Lovell-Badge, and Daniela Nebenius-Oosthuizen for discussions and help. This work was partly funded by a grant from the Volkswagen Foundation, Program on Conditional Mutagenesis. The work was initiated at the European Molecular Biology Laboratory (EMBL), Heidelberg.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The Red/ET DNA engineering technology is the subject of two issued patents authored by Drs. Zhang and Stewart. These patents are owned by EMBL which issued exclusive rights to GeneBridges GmbH, a company cofounded by Drs. Zhang and Stewart to develop the commercial implications of the patents. Dr. Zhang is CSO and Dr. Stewart chairperson of GeneBridges GmbH. Both are shareholders.
Rights and permissions
About this article
Cite this article
Testa, G., Zhang, Y., Vintersten, K. et al. Engineering the mouse genome with bacterial artificial chromosomes to create multipurpose alleles. Nat Biotechnol 21, 443–447 (2003). https://doi.org/10.1038/nbt804
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt804
This article is cited by
-
Generation and characterization of tissue-type plasminogen activator transgenic rats
Journal of Thrombosis and Thrombolysis (2018)
-
Beyond editing to writing large genomes
Nature Reviews Genetics (2017)
-
REPLACR-mutagenesis, a one-step method for site-directed mutagenesis by recombineering
Scientific Reports (2016)
-
Conditional knockout of Foxc2 gene in kidney: efficient generation of conditional alleles of single-exon gene by double-selection system
Mammalian Genome (2016)
-
A novel transchromosomic system: stable maintenance of an engineered Mb-sized human genomic fragment translocated to a mouse chromosome terminal region
Transgenic Research (2014)