Abstract
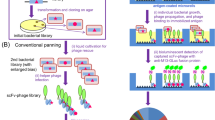
A nonimmune library of 109 human antibody scFv fragments has been cloned and expressed on the surface of yeast, and nanomolar-affinity scFvs routinely obtained by magnetic bead screening and flow-cytometric sorting. The yeast library can be amplified 1010-fold without measurable loss of clonal diversity, allowing its effectively indefinite expansion. The expression, stability, and antigen-binding properties of >50 isolated scFv clones were assessed directly on the yeast cell surface by immunofluorescent labeling and flow cytometry, obviating separate subcloning, expression, and purification steps and thereby expediting the isolation of novel affinity reagents. The ability to use multiplex library screening demonstrates the usefulness of this approach for high-throughput antibody isolation for proteomics applications.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Schweitzer, B. et al. Multiplexed protein profiling on microarrays by rolling-circle amplification. Nat. Biotechnol. 20, 359–365 (2002).
MacBeath, G. & Schreiber, S.L. Printing proteins as microarrays for high-throughput function determination. Science 289, 1760–1763 (2000).
Gura, T. Therapeutic antibodies: magic bullets hit the target. Nature 417, 584–586 (2002).
Clark, M. Antibody humanization: a case of the 'Emperor's new clothes'? Immunol. Today 21, 397–402 (2000).
Mendez, M.J. et al. Functional transplant of megabase human immunoglobulin loci recapitulates human antibody response in mice. Nat. Genet. 15, 146–156 (1997).
Winter, G., Griffiths, A.D., Hawkins, R.E. & Hoogenboom, H.R. Making antibodies by phage display technology. Annu. Rev. Immunol. 12, 433–455 (1994).
Marks, J.D. et al. Bypassing immunization. Human antibodies from V-gene libraries displayed on phage. J. Mol. Biol. 222, 581–597 (1991).
Sheets, M.D. et al. Efficient construction of a large nonimmune phage antibody library: the production of high-affinity human single-chain antibodies to protein antigens. Proc. Natl. Acad. Sci. USA 95, 6157–6162 (1998).
Vaughan, T.J. et al. Human antibodies with subnanomolar affinities isolated from a large nonimmunized phage display library. Nat. Biotechnol. 14, 309–314 (1996).
Knappik, A. et al. Fully synthetic human combinatorial antibody libraries (HuCAL) based on modular consensus frameworks and CDRs randomized with trinucleotides. J. Mol. Biol. 296, 57–86 (2000).
Hanes, J., Schaffitzel, C., Knappik, A. & Plückthun, A. Picomolar affinity antibodies from a fully synthetic naive library selected and evolved by ribosome display. Nat. Biotechnol. 18, 1287–1292 (2000).
Shusta, E.V., Holler, P.D., Kieke, M.C., Kranz, D.M. & Wittrup, K.D. Directed evolution of a stable scaffold for T-cell receptor engineering. Nat. Biotechnol. 18, 754–759 (2000).
Holler, P.D. et al. In vitro evolution of a T-cell receptor with high affinity for peptide/MHC. Proc. Natl. Acad. Sci. USA 97, 5387–5392 (2000).
Boder, E.T., Midelfort, K.S. & Wittrup, K.D. Directed evolution of antibody fragments with monovalent femtomolar antigen-binding affinity. Proc. Natl. Acad. Sci. USA 97, 10701–10705 (2000).
Daugherty, P.S., Iverson, B.L. & Georgiou, G. Flow cytometric screening of cell-based libraries. J. Immunol. Methods 243, 211–227 (2000).
VanAntwerp, J.J. & Wittrup, K.D. Fine affinity discrimination by yeast surface display and flow cytometry. Biotechnol. Prog. 16, 31–37 (2000).
Yeung, Y.A. & Wittrup, K.D. Quantitative screening of yeast surface-displayed polypeptide libraries by magnetic bead capture. Biotechnol. Prog. 18, 212–220 (2002).
Sblattero, D. & Bradbury, A. A definitive set of oligonucleotide primers for amplifying human V regions. Immunotechnology 3, 271–278 (1998).
Ignatovich, O., Tomlinson, I.M., Jones, P.T. & Winter, G. The creation of diversity in the human immunoglobulin V(λ) repertoire. J. Mol. Biol. 268, 69–77 (1997).
Foster, S.J., Brezinschek, H.P., Brezinschek, R.I. & Lipsky, P.E. Molecular mechanisms and selective influences that shape the κ-gene repertoire of IgM+ B cells. J. Clin. Invest. 99, 1614–1627 (1997).
Brezinschek, H.P. et al. Analysis of the human VH gene repertoire. Differential effects of selection and somatic hypermutation on human peripheral CD5+/IgM+ and CD5−/IgM+ B cells. J. Clin. Invest. 99, 2488–2501 (1997).
Ellgaard, L., Molinari, M. & Helenius, A. Setting the standards: quality control in the secretory pathway. Science 286, 1882–1888 (1999).
Field, C. & Schekman, R. Localized secretion of acid phosphatase reflects the pattern of cell surface growth in Saccharomyces cerevisiae. J. Cell Biol. 86, 123–128 (1980).
Shusta, E.V., Raines, R.T., Plückthun, A. & Wittrup, K.D. Increasing the secretory capacity of Saccharomyces cerevisiae for production of single-chain antibody fragments. Nat. Biotechnol. 16, 773–777 (1998).
Huang, W., McKevitt, M. & Palzkill, T. Use of the arabinose p(bad) promoter for tightly regulated display of proteins on bacteriophage. Gene 251, 187–197 (2000).
Krebber, A., Burmester, J. & Plückthun, A. Inclusion of an upstream transcriptional terminator in phage display vectors abolishes background expression of toxic fusions with coat protein g3p. Gene 178, 71–74 (1996).
Daugherty, P.S., Olsen, M.J., Iverson, B.L. & Georgiou, G. Development of an optimized expression system for the screening of antibody libraries displayed on the Escherichia coli surface. Protein Eng. 12, 613–621 (1999).
Lou, J. et al. Antibodies in haystacks: how selection strategy influences the outcome of selection from molecular diversity libraries. J. Immunol. Methods 253, 233–242 (2001).
Boder, E.T. & Wittrup, K.D. Yeast surface display for directed evolution of protein expression, affinity, and stability. Methods Enzymol. 328, 430–444 (2000).
Chen, G. et al. Isolation of high-affinity ligand-binding proteins by periplasmic expression with cytometric screening (PECS). Nat. Biotechnol. 19, 537–542 (2001).
Sblattero, D. & Bradbury, A. Exploiting recombination in single bacteria to make large phage antibody libraries. Nat. Biotechnol. 18, 75–80 (2000).
de Haard, H.J. et al. A large non-immunized human Fab fragment phage library that permits rapid isolation and kinetic analysis of high affinity antibodies. J. Biol. Chem. 274, 18218–18230 (1999).
Griffiths, A.D. et al. Isolation of high affinity human antibodies directly from large synthetic repertoires. EMBO J. 13, 3245–3260 (1994).
Soderlind, E. et al. Recombining germline-derived CDR sequences for creating diverse single-framework antibody libraries. Nat. Biotechnol. 18, 852–856 (2000).
Pini, A. et al. Design and use of a phage display library. Human antibodies with subnanomolar affinity against a marker of angiogenesis eluted from a two-dimensional gel. J. Biol. Chem. 273, 21769–21776 (1998).
Kieke, M.C. et al. High affinity T cell receptors from yeast display libraries block T cell activation by superantigens. J. Mol. Biol. 307, 1305–1315 (2001).
Llorente, M. et al. Natural human antibodies retrieved by phage display libraries from healthy donors: polyreactivity and recognition of human immunodeficiency virus type 1gp120 epitopes. Scand. J. Immunol. 50, 270–279 (1999).
Nieba, L., Honegger, A., Krebber, C. & Plückthun, A. Disrupting the hydrophobic patches at the antibody variable/constant domain interface: improved in vivo folding and physical characterization of an engineered scFv fragment. Protein Eng. 10, 435–444 (1997).
Bothmann, H. & Plückthun, A. Selection for a periplasmic factor improving phage display and functional periplasmic expression. Nat. Biotechnol. 16, 376–380 (1998).
de Bruin, R., Spelt, K., Mol, J., Koes, R. & Quattrocchio, F. Selection of high-affinity phage antibodies from phage display libraries. Nat. Biotechnol. 17, 397–399 (1999).
de Wildt, R.M., Mundy, C.R., Gorick, B.D. & Tomlinson, I.M. Antibody arrays for high-throughput screening of antibody–antigen interactions. Nat. Biotechnol. 18, 989–994 (2000).
Braun, P. et al. Proteome-scale purification of human proteins from bacteria. Proc. Natl. Acad. Sci. USA 99, 2654–2659. (2002).
Gavin, A.C. et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147 (2002).
Gietz, R.D., Schiestl, R.H., Willems, A.R. & Woods, R.A. Studies on the transformation of intact yeast cells by the LiAc/SS– DNA/PEG procedure. Yeast 11, 355–360 (1995).
Boder, E.T. & Wittrup, K.D. Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 15, 553–557 (1997).
Krebs, B. et al. High-throughput generation and engineering of recombinant human antibodies. J. Immunol. Methods 254, 67–84 (2001).
Acknowledgements
This work was funded in part by NSF BPEC, the Hereditary Disease Foundation, and by the Laboratory Directed Research and Development program of the US Department of Energy (DOE). The p53 phosphopeptides were provided by Ettore Appella, Shari Mazur, and Yuichiro Higashimoto (NCI). The authors are grateful for comments on the manuscript by David Kranz, Michael Roguska, and Eric Shusta.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
K.D.W. is an inventor listed on US patents 6423538, 6300065, and related filings that pertain to yeast surface display methodology, and receives a portion of any royalty income paid to the University of Illinois for licensing of these patents.
Rights and permissions
About this article
Cite this article
Feldhaus, M., Siegel, R., Opresko, L. et al. Flow-cytometric isolation of human antibodies from a nonimmune Saccharomyces cerevisiae surface display library. Nat Biotechnol 21, 163–170 (2003). https://doi.org/10.1038/nbt785
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt785
This article is cited by
-
A spike protein S2 antibody efficiently neutralizes the Omicron variant
Cellular & Molecular Immunology (2022)
-
A pandemic-enabled comparison of discovery platforms demonstrates a naïve antibody library can match the best immune-sourced antibodies
Nature Communications (2022)
-
Streamlined human antibody generation and optimization by exploiting designed immunoglobulin loci in a B cell line
Cellular & Molecular Immunology (2021)
-
Structural basis of ALC1/CHD1L autoinhibition and the mechanism of activation by the nucleosome
Nature Communications (2021)
-
Yeast display of MHC-II enables rapid identification of peptide ligands from protein antigens (RIPPA)
Cellular & Molecular Immunology (2021)