Abstract
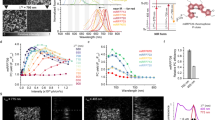
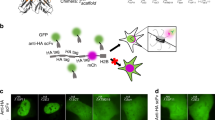
Photobleaching of green fluorescent protein (GFP) is a widely used approach for tracking the movement of subcellular structures and intracellular proteins1,2,3. Although photobleaching is a powerful technique, it does not allow direct tracking of an object's movement and velocity within a living cell. Direct tracking becomes possible only with the introduction of a photoactivated fluorescent marker. A number of previous studies have reported optically induced changes in the emission spectra of fluorescent proteins4,5,6,7. However, the ideal photoactivated fluorescent marker should be a nonfluorescent tag capable of “switching on” (i.e., becoming fluorescent) in response to irradiation by light of a particular wavelength, intensity, and duration. In this report, we generated a mutant of Anemonia sulcata chromoprotein asCP8. The mutant protein is capable of unique irreversible photoconversion from the nonfluorescent to a stable bright-red fluorescent form (“kindling”). This “kindling fluorescent protein” (KFP1) can be used for precise in vivo photolabeling to track the movements of cells, organelles, and proteins. We used KFP1 for in vivo cell labeling in mRNA microinjection assays to monitor Xenopus laevis embryo development and to track mitochondrial movement in mammalian cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
White, J. & Stelzer, E. Photobleaching GFP reveals protein dynamics inside live cells. Trends. Cell Biol. 9, 61–65 (1999).
Reits, E.A. & Neefjes, J.J. From fixed to FRAP: measuring protein mobility and activity in living cells. Nat. Cell Biol. 3, 145–147 (2001).
Lippincott-Schwartz, J., Snapp, E. & Kenworthy, A. Studying protein dynamics in living cells. Nat. Rev. Mol. Cell Biol. 2, 444–456 (2001).
Yokoe, H. & Meyer, T. Spatial dynamics of GFP-tagged proteins investigated by local fluorescence enhancement. Nat. Biotechnol. 14, 1252–1256 (1996).
Elowitz, M.B., Surette, M.G., Wolf, P., Stock, J. & Leibler, S. Photoactivation turns green fluorescent protein red. Curr. Biol. 7, 809–812 (1997).
Marchant, J.S., Stutzmann, G.E., Leissring, M.A., LaFerla, F.M., & Parker, I. Multiphoton-evoked color change of DsRed as an optical highlighter for cellular and subcellular labeling. Nat. Biotechnol. 19, 645–649 (2001).
Dunn, G.A., Dobbie, I.M., Monypenny, J., Holt, M.R. & Zicha, D. Fluorescence localization after photobleaching (FLAP): a new method for studying protein dynamics in living cells. J. Microsc. 205, 109–112 (2002).
Lukyanov, K.A. et al. Natural animal coloration can be determined by a nonfluorescent green fluorescent protein homolog. J. Biol. Chem. 275, 25879–25882 (2000).
Yang, F., Moss, L.G. & Phillips, G.N. The molecular structure of green fluorescent protein. Nat. Biotechnol. 14, 1246–1251 (1996).
Ormö, M. et al. Crystal structure of the Aequorea victoria green fluorescent protein. Science 273, 1392–1395 (1996).
Wall, M.A., Socolich, M. & Ranganathan, R. The structural basis for red fluorescence in the tetrameric GFP homolog DsRed. Nat. Struct. Biol. 7, 1133–1138 (2000).
Yarbrough, D., Wachter, R.M., Kallio, K., Matz, M.V. & Remington, S.J. Refined crystal structure of DsRed, a red fluorescent protein from coral, at 2.0 Å resolution. Proc. Natl. Acad. Sci. USA 98, 462–467 (2001).
Bulina, M., Chudakov, D., Mudrik, N. & Lukyanov, K. Interconversion of Anthozoa GFP-like fluorescent and non-fluorescent proteins by mutagenesis. BMC Biochem. 3, 7 (2002).
Yanushevich, Y.G. et al. A strategy for the generation of non-aggregating mutants of Anthozoa fluorescent proteins. FEBS Lett. 511, 11–14 (2002).
Keller, R. et al. Mechanisms of convergence and extension by cell intercalation. Phil. Trans. R. Soc. Lond. B Biol. Sci. 355, 897–922 (2000).
Condeelis, J.S., Wyckoff, J. & Segall, J.E. Imaging of cancer invasion and metastasis using green fluorescent protein. Eur. J. Cancer 36, 1671–1680 (2000).
Vajkoczy, P., Ullrich, A. & Menger, M.D. Intravital fluorescence videomicroscopy to study tumor angiogenesis and microcirculation. Neoplasia 2, 53–61 (2000).
Parish, C.R. Fluorescent dyes for lymphocyte migration and proliferation studies. Immunol. Cell Biol. 77, 499–508 (1999).
Campbell, R.E. et al. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 11, 7877–7882 (2002).
Matz, M.V. et al. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat. Biotechnol. 17, 969–973 (1999).
Patterson, G.H. & Lippincott-Schwartz, J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science 13, 1873–1877 (2002).
Ando, R., Hama, H., Yamamoto-Hino, M., Mizuno, H. & Miyawaki, A. An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc. Natl. Acad. Sci. USA 99, 12651–12656 (2002).
Ho, S.N., Hunt, H.D., Horton, R.M., Pullen, J.K. & Pease, L.R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59 (1989).
Nieuwkoop, P.D. & Faber, J. Normal Table of Xenopus laevis (Daudin) (North-Holland, Amsterdam, 1967).
Patterson, G., Day, R. & N. Piston D. Fluorescent protein spectra. J. Cell Sci. 114, 837–838 (2001).
Acknowledgements
The authors thank A.V. Feofanov (Shemiakin and Ovchinnikov Institute of Bioorganic Chemistry RAS) for valuable help in intracellular fluorescence kindling technique development. This work was supported by the Russian Foundation for Basic Research (grant 01-04-49037), the Russian Foundation for Support of Domestic Science grant to S.A.L., and by grants from HHMI (55000344), FIRCA, and CRDF (RB1-2406-MO-02) to A.G.Z.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Chudakov, D., Belousov, V., Zaraisky, A. et al. Kindling fluorescent proteins for precise in vivo photolabeling. Nat Biotechnol 21, 191–194 (2003). https://doi.org/10.1038/nbt778
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt778
This article is cited by
-
Targeting mitochondrial shape: at the heart of cardioprotection
Basic Research in Cardiology (2023)
-
Chromophore reduction plus reversible photobleaching: how the mKate2 “photoconversion” works
Photochemical & Photobiological Sciences (2021)
-
Fast reversibly photoswitching red fluorescent proteins for live-cell RESOLFT nanoscopy
Nature Methods (2018)
-
Role of green fluorescent proteins and their variants in development of FRET-based sensors
Journal of Biosciences (2018)
-
Choosing proper fluorescent dyes, proteins, and imaging techniques to study mitochondrial dynamics in mammalian cells
Biophysics Reports (2017)