Abstract
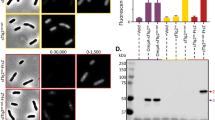
We demonstrate the use of Propionibacterium freudenreichii uroporphyrinogen III methyltransferase (cobA) as a reporter of gene expression in Escherichia coli, fission yeast, and mammalian cells. Overexpression of cobA in cells resulted in bright red fluorescence that was visualized with standard fluorescence microscopy and fluorescence-activated cell sorting analysis at the single-cell level. As with green fluorescent protein (GFP), no addition of exogenous substrate was required. When expressed in Chinese hamster ovary cells from a bicistronic transcript, cobA and GFP gave rise to fluorescence signals of similar intensity. The bright red fluorescence generated by the cobA reporter promises a better signal-to-noise ratio than blue and green fluorescent reporter systems, as autofluorescence and light scattering of cells, media, and materials are reduced in the red wavelengths.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
An, G., Hidaka, K. & Siminovitch, L. Expression of bacterial beta-galactosidase in animal cells. Mol. Cell. Biol. 2, 1628– 1632 (1982).
Gorman, C.M., Moffat, L.F. & Howard, B.H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol. Cell. Biol. 2, 1044–1051 (1982).
Gould, S.J. & Subramani, S. Firefly luciferase as a tool in molecular and cell biology. Anal. Biochem. 175, 5–13 (1988).
Berger, J., Hauber, J., Hauber, R., Geiger, R. & Cullen, B.R. Secreted placental alkaline phosphatase: a powerful new quantitative indicator of gene expression in eukaryotic cells. Gene 66, 1–10 ( 1988).
Yoon, K., Thiede, M.A. & Rodan, G.A. Alkaline phosphatase as a reporter enzyme. Gene 66, 11–17 ( 1988).
Chalfie, M., Tu, Y., Euskirchen, G., Ward, W.W. & Prasher, D.C. Green fluorescent protein as a marker for gene expression. Science 263, 802–805 (1994).
Millar, A.J., Carre, I.A., Strayer, C.A., Chua, N.-H. & Kay, S.A. Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science 267, 1161–1163 (1995).
Millar, A.J., Straume, M., Chory, J., Chua, N.-H. & Kay, S.A. The regulation of phototransduction pathways in Arabidopsis . Science 267, 1163– 1166 (1995).
Plautz, J.D. et al. Green fluorescent protein and its derivatives as versatile markers for gene expression in living Drosophila melanogaster, plant and mammalian cells. Gene 173, 83–87 (1996).
Zlokarnik, G. et al. Quantitation of transcription and clonal selection of single living cells with beta-lactamase as reporter. Science 279, 84–88 (1998).
Warren, M.J., Roessner, C.A., Santander, P.J. & Scott, A.I. The Escherichia coli cysG gene encodes S-adenosylmethionine-dependent uroporphyrinogen III methylase. Biochem. J. 265, 725–729 (1990).
Sattler, I. et al. Cloning, sequencing, and expression of the uroporphyrinogen III methyltransferase cobA gene of Propionibacterium freudenreichii (shermanii). J. Bacteriol. 177, 1564–1569 (1995).
Roessner, C.A. & Scott, A.I. Fluorescence-based method for selection of recombinant plasmids. Biotechniques 19, 760–764 (1995).
Scott, A.I. Recent studies of enzymically controlled steps in B12 biosynthesis. Ciba Found Symp. 180, 285–303 (1994).
Stüber, D., Matile, H. & Garotta, G. in Immunological methods Vol. IV I. Leftovits & B. Pernis, Eds. Pe121–152 (Academic Press, Orlando 1990).
Maundrell, K. nmt1 of fission yeast. J. Biol. Chem. 265, 10857–10864 (1990).
Kozak, M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44, 283–292 (1986).
No, D., Yao, T.P. & Evans, R.M. Ecdysone-inducible gene expression in mammalian cells and transgenic mice. Proc. Natl. Acad. Sci. USA 93, 3346–3351 (1996).
Dirks, W., Wirth, H. & Hauser, H. Dicistronic transcription units for gene expression in mammalian cells. Gene 128, 247– 249 (1993).
Cormack, B.P., Valdivia, R.H. & Falkow, S. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173, 33–38 (1996).
Leustek, T. et al. Siroheme biosynthesis in higher plants. Analysis of an S-adenosyl-L-methionine-dependent uroporphyrinogen III methyltransferase from Arabidopsis thaliana. J. Biol. Chem. 272, 2744–2752 (1997).
Sakakibara, H., Takei, K. & Sugijama, T. Isolation and characterization of a cDNA that encodes maize uroporphyrinogen III methyltransferase, an enzyme involved in the synthesis of siroheme, which is a prosthetic group of nitrite reductase. Plant J. 10, 883–892 ( 1996).
Raux, E., McVeigh, T., Peters, S.E., Leustek, T. & Warren, M. The role of Saccharomyces cerevisiae Met1p and Met8p in sirohaem and cobalamin biosynthesis. Biochem.J. 338, 701–708 ( 1999).
Chou, P.L. et al. Reddish Escherichia coli cells caused by overproduction of Bacillus stearothermophilus uroporphyrinogen III methylase: cloning, sequencing, and expression of the gene. Biosci. Biotechnol. Biochem. 59, 1817–1824 (1995).
Matz, M.V. et al. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat. Biotechnol. 17, 969–973 (1999).
Tsien, R.Y. Rosy dawn for fluorescent proteins. Nat. Biotechnol. 17, 956–957 (1999).
Moreno, S., Klar, A. & Nurse, P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795– 823 (1991).
Acknowledgements
We would like to thank Dr. Charles A. Roessner for the gift of pISA417; Adriana Missano, Nicole Holzwarth, Nina Barth, and Ashley Hayes for technical assistance; and Dr. Francis Mueller and E. Kunitz for help using the spectrophotometer.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wildt, S., Deuschle, U. cobA, a red fluorescent transcriptional reporter for Escherichia coli, yeast, and mammalian cells. Nat Biotechnol 17, 1175–1178 (1999). https://doi.org/10.1038/70713
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/70713
This article is cited by
-
Cloning and heterologous expression of Lactobacillus reuteri uroporphyrinogen III synthase/methyltransferase gene (cobA/hemD): preliminary characterization
Biotechnology Letters (2011)
-
Creating new fluorescent probes for cell biology
Nature Reviews Molecular Cell Biology (2002)