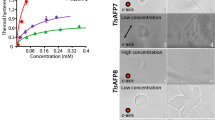
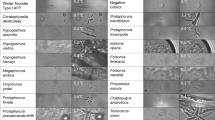
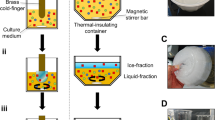
Abstract
Antifreeze proteins (AFPs) are found in cold-adapted organisms and have the unusual ability to bind to and inhibit the growth of ice crystals. However, the underlying molecular basis of their ice-binding activity is unclear because of the difficulty of studying the AFP-ice interaction directly and the lack of a common motif, domain or fold among different AFPs. We have formulated a generic ice-binding model and incorporated it into a physicochemical pattern-recognition algorithm. It successfully recognizes ice-binding surfaces for a diverse range of AFPs, and clearly discriminates AFPs from other structures in the Protein Data Bank1. The algorithm was used to identify a novel AFP from winter rye, and the antifreeze activity of this protein was subsequently confirmed. The presence of a common and distinct physicochemical pattern provides a structural basis for unifying AFPs from fish, insects and plants.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Berman, H.M. et al. The Protein Data Bank. Nucleic Acids Res. 28, 235–242 (2000).
Duman, J. & DeVries, A. Freezing resistance in winter flounder Pseudopleuronectes americanus . Nature 247, 237–238 (1974).
Knight, C.A. Adding to the antifreeze agenda. Nature 406, 249–251 (2000).
Jia, Z. & Davies, P.L. Antifreeze proteins: an unusual receptor-ligand interaction. Trends Biochem. Sci. 27, 101–106 (2002).
Jorov, A., Zhorov, B.S. & Yang, D.S. Theoretical study of interaction of winter flounder antifreeze protein with ice. Protein Sci. 13, 1524–1537 (2004).
Harding, M.M., Ward, L.G. & Haymet, A.D. Type I 'antifreeze' proteins. Structure-activity studies and mechanisms of ice growth inhibition. Eur. J. Biochem. 264, 653–665 (1999).
Yang, D.S. et al. Identification of the ice-binding surface on a type III antifreeze protein with a “flatness function” algorithm. Biophys. J. 74, 2142–2151 (1998).
DeLuca, C.I., Davies, P.L., Ye, Q. & Jia, Z. The effects of steric mutations on the structure of type III antifreeze protein and its interaction with ice. J. Mol. Biol. 275, 515–525 (1998).
Gallagher, K.R. & Sharp, K.A. Analysis of thermal hysteresis protein hydration using the random network model. Biophys. Chem. 105, 195–209 (2003).
Antson, A.A. et al. Understanding the mechanism of ice binding by type III antifreeze proteins. J. Mol. Biol. 305, 875–889 (2001).
Graether, S.P. & Sykes, B.D. Cold survival in freeze-intolerant insects: the structure and function of beta-helical antifreeze proteins. Eur. J. Biochem. 271, 3285–3296 (2004).
Chao, H., Sonnichsen, F.D., DeLuca, C.I., Sykes, B.D. & Davies, P.L. Structure-function relationship in the globular type III antifreeze protein: identification of a cluster of surface residues required for binding to ice. Protein Sci. 3, 1760–1769 (1994).
Graether, S.P. et al. Quantitative and qualitative analysis of type III antifreeze protein structure and function. J. Biol. Chem. 274, 11842–11847 (1999).
Jia, Z., DeLuca, C.I., Chao, H. & Davies, P.L. Structural basis for the binding of a globular antifreeze protein to ice. Nature 384, 285–288 (1996).
Fernandez-Recio, J., Totrov, M., Skorodumov, C. & Abagyan, R. Optimal docking area: a new method for predicting protein-protein interaction sites. Proteins 58, 134–143 (2005).
Hincha, D.K. et al. Cabbage cryoprotectin is a member of the nonspecific plant lipid transfer protein gene family. Plant Physiol. 125, 835–846 (2001).
DeVries, A.L. Antifreeze glycopeptides and peptides: interactions with ice and water. Methods Enzymol. 127, 293–303 (1986).
Griffith, M. & Yaish, M.W. Antifreeze proteins in overwintering plants: a tale of two activities. Trends Plant Sci. 9, 399–405 (2004).
Hon, W.C., Griffith, M., Chong, P. & Yang, D. Extraction and isolation of antifreeze proteins from winter rye (Secale cereale L.) leaves. Plant Physiol. 104, 971–980 (1994).
Snider, C., Hsiang, T., Zhao, G. & Griffith, M. Role of ice nucleation and antifreeze activities in pathogenesis and growth of snow molds. Phytopathology 90, 354–361 (2000).
Gauthier, S.Y., Kay, C.M., Sykes, B.D., Walker, V.K. & Davies, P.L. Disulfide bond mapping and structural characterization of spruce budworm antifreeze protein. Eur. J. Biochem. 258, 445–453 (1998).
Sieg, F., Schroder, W., Schmitt, J.M. & Hincha, D.K. Purification and characterization of a cryoprotective protein (cryoprotectin) from the leaves of cold-acclimated cabbage. Plant Physiol. 111, 215–221 (1996).
McConkey, B.J., Sobolev, V. & Edelman, M. Quantification of protein surfaces, volumes and atom-atom contacts using a constrained Voronoi procedure. Bioinformatics 18, 1365–1373 (2002).
Madura, J.D., Baran, K. & Wierzbicki, A. Molecular recognition and binding of thermal hysteresis proteins to ice. J. Mol. Recognit. 13, 101–113 (2000).
Wang, G. & Dunbrack, R.L., Jr. PISCES: a protein sequence culling server. Bioinformatics 19, 1589–1591 (2003).
Kelley, L.A. & Sutcliffe, M.J. OLDERADO: on-line database of ensemble representatives and domains. Protein Sci. 6, 2628–2630 (1997).
Schwede, T., Kopp, J., Guex, N. & Peitsch, M.C. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 31, 3381–3385 (2003).
Guex, N. & Peitsch, M.C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723 (1997).
Hooft, R.W., Vriend, G., Sander, C. & Abola, E.E . Errors in protein structures. Nature 381, 272 (1996).
Bravo, L.A. & Griffith, M. Characterization of antifreeze activity in Antarctic plants. J. Exp. Bot. 56, 1189–1196 (2005).
Acknowledgements
We would like to dedicate this publication to the memory of Marilyn Griffith. This work was supported by grants from the National Science and Engineering Research Council (NSERC) to B.J.M. and M.G., and we acknowledge the Killam Research Fellowship Program on behalf of M.G. We thank Barbara Moffatt for providing T. salsuginea sequences and Bernard Duncker for comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Ordered surface carbons detected in known AFPs. (PDF 1064 kb)
Supplementary Fig. 2
Correlation between type III mutant thermal hysteresis values and algorithm score. (PDF 64 kb)
Supplementary Fig. 3
Algorithm for prediction of ordered surface carbons. (PDF 68 kb)
Supplementary Table 1
Residues contributing predicted ordered surface carbons. (PDF 47 kb)
Supplementary Table 2
Comparison of algorithm performance with individual components omitted. (PDF 37 kb)
Rights and permissions
About this article
Cite this article
Doxey, A., Yaish, M., Griffith, M. et al. Ordered surface carbons distinguish antifreeze proteins and their ice-binding regions. Nat Biotechnol 24, 852–855 (2006). https://doi.org/10.1038/nbt1224
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt1224
This article is cited by
-
Soy β-Conglycinin−Curcumin Nanocomplexes for Enrichment of Clear Beverages
Food Biophysics (2015)
-
Freezing tolerance in Norway spruce, the potential role of pathogenesis-related proteins
Acta Physiologiae Plantarum (2015)
-
Diverse chitinases are invoked during the activity-dormancy transition in spruce
Tree Genetics & Genomes (2015)
-
Characterization of Afp1, an antifreeze protein from the psychrophilic yeast Glaciozyma antarctica PI12
Extremophiles (2013)
-
RETRACTED ARTICLE: Isolation and Functional Characterization of an Antifreeze Protein Gene, TaAFPIII, from Wheat (Triticum aestivum)
Plant Molecular Biology Reporter (2012)