Abstract
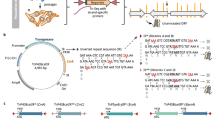
Cloning of the entire set of an organism's protein-coding open reading frames (ORFs), or 'ORFeome', is a means of connecting the genome to downstream 'omics' applications. Here we report a proteome-scale study of the fission yeast Schizosaccharomyces pombe based on cloning of the ORFeome. Taking advantage of a recombination-based cloning system, we obtained 4,910 ORFs in a form that is readily usable in various analyses. First, we evaluated ORF prediction in the fission yeast genome project by expressing each ORF tagged at the 3′ terminus. Next, we determined the localization of 4,431 proteins, corresponding to ∼90% of the fission yeast proteome, by tagging each ORF with the yellow fluorescent protein. Furthermore, using leptomycin B, an inhibitor of the nuclear export protein Crm1, we identified 285 proteins whose localization is regulated by Crm1.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Huh, W.K. et al. Global analysis of protein localization in budding yeast. Nature 425, 686–691 (2003).
Reboul, J. et al. C. elegans ORFeome version 1.1: experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat. Genet. 34, 35–41 (2003).
Wood, V. et al. The genome sequence of Schizosaccharomyces pombe . Nature 415, 871–880 (2002).
Matsuyama, A. et al. pDUAL, a multipurpose, multicopy vector capable of chromosomal integration in fission yeast. Yeast 21, 1289–1305 (2004).
Maundrell, K. nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J. Biol. Chem. 265, 10857–10864 (1990).
Poetz, O. et al. Protein microarrays: catching the proteome. Mech. Ageing Dev. 126, 161–170 (2005).
Tsien, R.Y. The green fluorescent protein. Annu. Rev. Biochem. 67, 509–544 (1998).
Paoletti, A. et al. Fission yeast cdc31p is a component of the half-bridge and controls SPB duplication. Mol. Biol. Cell 14, 2793–2808 (2003).
D'Alessio, C., Trombetta, E.S. & Parodi, A.J. Nucleoside diphosphatase and glycosyltransferase activities can localize to different subcellular compartments in Schizosaccharomyces pombe . J. Biol. Chem. 278, 22379–22387 (2003).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 (2000).
Burgess, D.R. & Chang, F. Site selection for the cleavage furrow at cytokinesis. Trends Cell Biol. 15, 156–162 (2005).
Zimber, A., Nguyen, Q.D. & Gespach, C. Nuclear bodies and compartments: functional roles and cellular signalling in health and disease. Cell. Signal. 16, 1085–1104 (2004).
Ullman, K.S., Powers, M.A. & Forbes, D.J. Nuclear export receptors: from importin to exportin. Cell 90, 967–970 (1997).
Bogerd, H.P., Fridell, R.A., Benson, R.E., Hua, J. & Cullen, B.R. Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol. Cell. Biol. 16, 4207–4214 (1996).
Kudo, N., Taoka, H., Toda, T., Yoshida, M. & Horinouchi, S. A novel nuclear export signal sensitive to oxidative stress in the fission yeast transcription factor Pap1. J. Biol. Chem. 274, 15151–15158 (1999).
Hoshino, H. et al. Oxidative stress abolishes leptomycin B-sensitive nuclear export of transcription repressor Bach2 that counteracts activation of Maf recognition element. J. Biol. Chem. 275, 15370–15376 (2000).
Kudo, N. et al. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell Res. 242, 540–547 (1998).
Wolff, B., Sanglier, J.J. & Wang, Y. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem. Biol. 4, 139–147 (1997).
Kudo, N. et al. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. USA 96, 9112–9117 (1999).
Nguyen, A.N., Ikner, A.D., Shiozaki, M., Warren, S.M. & Shiozaki, K. Cytoplasmic localization of Wis1 MAPKK by nuclear export signal is important for nuclear targeting of Spc1/Sty1 MAPK in fission yeast. Mol. Biol. Cell 13, 2651–2663 (2002).
Qin, J., Kang, W., Leung, B. & McLeod, M. Ste11p, a high-mobility-group box DNA-binding protein, undergoes pheromone- and nutrient-regulated nuclear-cytoplasmic shuttling. Mol. Cell. Biol. 23, 3253–3264 (2003).
Sutani, T. et al. Fission yeast condensin complex: essential roles of non-SMC subunits for condensation and Cdc2 phosphorylation of Cut3/SMC4. Genes Dev. 13, 2271–2283 (1999).
Paoletti, A. & Chang, F. Analysis of mid1p, a protein required for placement of the cell division site, reveals a link between the nucleus and the cell surface in fission yeast. Mol. Biol. Cell 11, 2757–2773 (2000).
Sipiczki, M. Where does fission yeast sit on the tree of life? Genome Biol. 1 REVIEWS1011 (2000).
Umeda, M., Izaddoost, S., Cushman, I., Moore, M.S. & Sazer, S. The fission yeast Schizosaccharomyces pombe has two importin-α, Imp1p and Cut15p, which have common and unique functions in nucleocytoplasmic transport and cell cycle progression. Genetics 171, 7–21 (2005).
Moreno, S., Klar, A. & Nurse, P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe . Methods Enzymol. 194, 795–823 (1991).
Matsuyama, A., Yabana, N., Watanabe, Y. & Yamamoto, M. Schizosaccharomyces pombe Ste7p is required for both promotion and withholding of the entry to meiosis. Genetics 155, 539–549 (2000).
Kushnirov, V.V. Rapid and reliable protein extraction from yeast. Yeast 16, 857–860 (2000).
Elder, R.T., Loh, E.Y. & Davis, R.W. RNA from the yeast transposable element Ty1 has both ends in the direct repeats, a structure similar to retrovirus RNA. Proc. Natl. Acad. Sci. USA 80, 2432–2436 (1983).
Csank, C. et al. Three yeast proteome databases: YPD, PombePD, and CalPD (MycoPathPD). Methods Enzymol. 350, 347–373 (2002).
Acknowledgements
We thank Megumi Takase, Kunio Watanabe and Ryuji Yoshinari for technical assistance and the members of Chemical Genetics Laboratory, RIKEN, for discussions. We also thank Daniel McCollum for critical reading of the manuscript, and Jürg Bähler for useful comments. This work was supported in part by the CREST Research Project, Japan Science and Technology Agency, The Strategic Research Programs for R&D, RIKEN and a Grant-in-Aid for Scientific Research on Priority Area 'Cancer' from The Ministry of Education, Culture, Sports, Science and Technology, Japan.
Author information
Authors and Affiliations
Contributions
The order of listing of the authors A.M., R.A. and Y.Y. in no way reflects their relative contribution to this work. M.Y. is responsible for the project planning and experimental design with support from Y.H. and S.H.; A.M., Y.Y. and A.H. carried out cloning and sequencing; R.A., Y.Y. and M.H. collected the localizome data; R.A. and A.K. carried out the chemical genetic screen; S.S. and Y.K. generated plasmids and transformants; and A.M. and A.S. carried out reverse array and statistic analyses.
Note: Supplementary information is available on the Nature Biotechnology website.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Distribution of the protein expression levels. (PDF 735 kb)
Supplementary Fig. 2
Comparison of the fission yeast localizome with the GO_cellular component categories. (PDF 873 kb)
Supplementary Fig. 3
Distribution pattern of proteins in cellular compartments in budding yeast. (PDF 831 kb)
Supplementary Table 1
Summary of S. pombe expression library construction. (PDF 75 kb)
Supplementary Table 2
ORFs that showed disagreement with the existing genome database. (PDF 77 kb)
Supplementary Table 3
Functional classification of 'non-SPB' nuclear dots proteins. (PDF 62 kb)
Supplementary Table 4
Screening of Crm1-dependently transported proteins. (PDF 68 kb)
Supplementary Table 5
Functional classification of Crm1-dependently transported proteins. (PDF 96 kb)
Supplementary Table 6
Sequence information and localizome data for the entry clones. (XLS 1736 kb)
Rights and permissions
About this article
Cite this article
Matsuyama, A., Arai, R., Yashiroda, Y. et al. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol 24, 841–847 (2006). https://doi.org/10.1038/nbt1222
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt1222
This article is cited by
-
A set of vectors and strains for chromosomal integration in fission yeast
Scientific Reports (2023)
-
Large-scale identification of genes involved in septal pore plugging in multicellular fungi
Nature Communications (2023)
-
Genome-wide subcellular protein map for the flagellate parasite Trypanosoma brucei
Nature Microbiology (2023)
-
Discovery of a novel marker for human granulocytes and tissue macrophages: RTL1 revisited
Cell and Tissue Research (2023)
-
Core control principles of the eukaryotic cell cycle
Nature (2022)