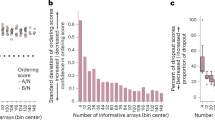
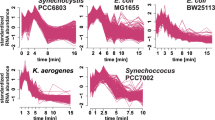
Abstract
We have developed a high-resolution “genome array” for the study of gene expression and regulation in Escherichia coli. This array contains on average one 25-mer oligonucleotide probe per 30 base pairs over the entire genome, with one every 6 bases for the intergenic regions and every 60 bases for the 4,290 open reading frames (ORFs). Twofold concentration differences can be detected at levels as low as 0.2 messenger RNA (mRNA) copies per cell, and differences can be seen over a dynamic range of three orders of magnitude. In rich medium we detected transcripts for 97% and 87% of the ORFs in stationary and log phases, respectively. We found that 1,529 transcripts were differentially expressed under these conditions. As expected, genes involved in translation were expressed at higher levels in log phase, whereas many genes known to be involved in the starvation response were expressed at higher levels in stationary phase. Many previously unrecognized growth phase-regulated genes were identified, such as a putative receptor (b0836) and a 30S ribosomal protein subunit (S22), both of which are highly upregulated in stationary phase. Transcription of between 3,000 and 4,000 predicted ORFs was observed from the antisense strand, indicating that most of the genome is transcribed at a detectable level. Examples are also presented for high-resolution array analysis of transcript start and stop sites and RNA secondary structure.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
de Saizieu, A. et al. Bacterial transcript imaging by hybridization of total RNA to oligonucleotide arrays. Nat. Biotechnol. 16, 45– 48 (1998).
Lockhart, D.J. et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 14, 1675– 1680 (1996).
Wodicka, L. et al. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat. Biotechnol. 15, 1359–1367 (1997).
Lee, C.K. et al. Gene expression profile of aging and its retardation by caloric restriction . Science 285, 1390–1393 (1999).
Zhu, H. et al. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95, 14470–14475 (1998).
Wen, X. et al. Large-scale temporal gene expression mapping of central nervous system development . Proc. Natl. Acad. Sci. USA 95, 334– 339 (1998).
Roth, F.P. et al. Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat. Biotechnol. 16, 939–945 (1998).
Tavazoie, S. et al. Systematic determination of genetic network architecture. Nat. Genet. 22, 281–285 (1999).
Eisen, M.B. et al. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95, 14863– 14868 (1998).
Tamayo, P. et al. Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc. Natl. Acad. Sci. USA 96, 2907–2912 ( 1999).
Richmond, C.S. et al. Genome-wide expression profiling in Escherichia coli K-12 . Nucleic Acids Res. 27, 3821– 3835 (1999).
Tao, H. et al. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J. Bacteriol. 181, 6425–6440 (1999).
Chuang, S.E., Daniels, D.L. & Blattner, F.R. Global regulation of gene expression in Escherichia coli. J. Bacteriol. 175, 2026– 2036 (1993).
Pease, A.C. et al. Light-generated oligonucleotide arrays for rapid DNA sequence analysis . Proc. Natl. Acad. Sci. USA 91, 5022– 5026 (1994).
Blattner, F.R. et al. The complete genome sequence of Escherichia coli K-12. Science 277, 1453–1474 ( 1997).
Hengge-Aronis, R. Regulation of gene expression during entry into stationary phase. In Escherichia coli and Salmonella: cellular and molecular biology. (eds Neidhardt, F.C. et al.) 1497–1512 (ASM Press, Washington D.C.; 1996).
Yuzawa, H. et al. Heat induction of sigma 32 synthesis mediated by mRNA secondary structure: a primary step of the heat shock response in Escherichia coli. Nucleic Acids Res. 21, 5449–5455 (1993).
Lange, R. & Hengge-Aronis, R. The cellular concentration of the sigma S subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, and protein stability. Genes Dev. 8, 1600– 1612 (1994).
E. coli genome project. http://www.genome.wisc.edu
Link, A.J., Robison, K. & Church, G.M. Comparing the predicted and observed properties of proteins encoded in the genome of Escherichia coli K-12. Electrophoresis 18, 1259–1313 (1997).
Aach, J., Rindone, W. & Church, G.M. Systematic management and analysis of yeast gene expression data. Genome Res. 10, 431– 445 (2000).
ExpressDB. http://arep.med.harvard.edu/cgi-bin/ExpressDBecoli/EXDStart
Liu, M.Y. et al. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J. Biol. Chem. 272, 17502–17510 (1997).
Southern, E.M., Milner, N. & Mir, K.U. Discovering antisense reagents by hybridization of RNA to oligonucleotide arrays. Ciba Found. Symp. 209 , 38–44 (1997).
Chen, L.H. et al. Structure and function of a bacterial mRNA stabilizer: analysis of the 5′ untranslated region of ompA mRNA. J. Bacteriol. 173, 4578–4586 (1991).
SantaLucia, J., Jr. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. USA 95, 1460–1465 ( 1998).
mfold. http://mfold2.wustl.edu/~mfold/dna/form1.cgi
Mir, K.U. & Southern, E.M. Determining the influence of structure on hybridization using oligonucleotide arrays. Nat. Biotechnol. 17, 788–792 (1999).
Taljanidisz, J., Karnik, P. & Sarkar, N. Messenger ribonucleic acid for the lipoprotein of the Escherichia coli outer membrane is polyadenylated. J. Mol. Biol. 193, 507–515 (1987).
Cao, G.J. & N. Sarkar . Poly(A) RNA in Escherichia coli: nucleotide sequence at the junction of the lpp transcript and the polyadenylate moiety. Proc. Natl. Acad. Sci. USA 89, 7546–7550 (1992).
Portier, C. & Regnier, P. Expression of the rpsO and pnp genes: structural analysis of a DNA fragment carrying their control regions. Nucleic Acids Res. 12, 6091–6102 (1984).
Portier, C. et al. The first step in the functional inactivation of the Escherichia coli polynucleotide phosphorylase messenger is a ribonuclease III processing at the 5′ end. EMBO J. 6, 2165– 2170 (1987).
Regnier, P. & Hajnsdorf, E. Decay of mRNA encoding ribosomal protein S15 of Escherichia coli is initiated by an RNase E-dependent endonucleolytic cleavage that removes the 3′ stabilizing stem and loop structure. J. Mol. Biol. 217, 283–292 (1991).
Pennisi, E. Are sequencers ready to 'annotate' the human genome? Science 287, 2183 (2000).
DeRisi, J. et al. Use of a cDNA microarray to analyse gene expression patterns in human cancer . Nat. Genet. 14, 457–460 (1996).
DeRisi, J.L., Iyer, V.R. & Brown, P.O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278, 680–686 (1997).
Tavazoie, S. & Church, G.M. Quantitative whole-genome analysis of DNA–protein interactions by in vivo methylase protection in E. coli. Nat. Biotechnol. 16, 566–571 (1998).
Dedon, P.C. et al. A simplified formaldehyde fixation and immunoprecipitation technique for studying protein–DNA interactions. Anal. Biochem. 197, 83–90 (1991).
Orlando, V. & Paro, R. Mapping Polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin . Cell 75, 1187–1198 (1993).
Bulyk, M.L. et al. Quantifying DNA–protein interactions by double-stranded DNA arrays. Nat. Biotechnol. 17, 573– 577 (1999).
Winzeler, E.A. et al. Whole genome genetic-typing in yeast using high-density oligonucleotide arrays. Parasitology 118, S73– S80 (1999).
Gingeras, T.R. et al. Simultaneous genotyping and species identification using hybridization pattern recognition analysis of generic Mycobacterium DNA arrays. Genome Res. 8, 435–448 ( 1998).
Neidhardt, F.C., Ingraham, J.L. & Schaechter, M. Physiology of the bacterial cell: a molecular approach. (Sinauer Associates, Sunderland, MA; 1990 ).
Acknowledgements
We thank J. Edwards for improvements to the labeling protocol, D. Janse for sharing unpublished data, A. Derti and A. Petti for bioinformatics contributions, F. Lam for help with the fermentor, M. Mittmann for array design, P. Juels for impeccable computer tech support, W. Rindone and J. Aach for expression database support, B. Cohen, R. Mitra, M. Bulyk, P. Estep, M. Steffen, and the rest of the Church lab for the many helpful discussions and encouragement that made this work possible. We also thank the reviewers for significant improvements to the manuscript. This work was supported by grants from Aventis Pharma, Lipper Foundation, DOE, and NSF.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Selinger, D., Cheung, K., Mei, R. et al. RNA expression analysis using a 30 base pair resolution Escherichia coli genome array. Nat Biotechnol 18, 1262–1268 (2000). https://doi.org/10.1038/82367
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/82367
This article is cited by
-
An electrochemical DNA biosensor based gold-thiolate conjugation utilizing ruthenium complex [Ru(dppz)2(qtpy)]Cl2
Microsystem Technologies (2017)
-
Au decorated ZnO thin film: application to DNA sensing
Microsystem Technologies (2016)
-
quantro: a data-driven approach to guide the choice of an appropriate normalization method
Genome Biology (2015)
-
Colony-live — a high-throughput method for measuring microbial colony growth kinetics— reveals diverse growth effects of gene knockouts in Escherichia coli
BMC Microbiology (2014)
-
Pervasive transcription: illuminating the dark matter of bacterial transcriptomes
Nature Reviews Microbiology (2014)