Abstract
The use of transgenic crops has generated concerns about transgene movement to unintended hosts and the associated ecological consequences. Moreover, the in-field monitoring of transgene expression is of practical concern (e.g., the underexpression of an herbicide tolerance gene in crop plants that are due to be sprayed with herbicide). A solution to these potential problems is to monitor the presence and expression of an agronomically important gene by linking it to a marker gene, such as GFP. Here we show that GFP fluorescence can indicate expression of the Bacillus thuringiensus cry1Ac gene when co-introduced into tobacco and oilseed rape, as demonstrated by insect bioassays and western blot analysis. Furthermore we conducted two seasons of field experiments to characterize the performance of three different GFP genes in transgenic tobacco. The best gene tested was mGFP5er, a mutagenized GFP gene that is targeted to the endoplasmic reticulum. We also demonstrated that host plants synthesizing GFP in the field suffered no fitness costs.
Similar content being viewed by others
Main
The use of transgenic plants in agriculture has caused concern about the possible introgression of transgenes into wild relatives through hybridization and backcrossing1, which may increase the fitness of wild relatives allowing them to proliferate in that environment1,2. The apparent risk of transgene escape warrants the development of gene monitoring systems that, ideally, are universally applicable and suitable for use in the field. Detection of antibiotic- and herbicide-resistance genes or scorable marker genes is not suitable for commercial-scale agriculture, because it requires destructive plant tissue sampling or time-consuming assays. A more desirable system would allow detection of transgenes and their expression in real time and on live plants.
Green fluorescent protein (GFP) would be suitable as an in vivo marker for monitoring transgenic plants, because it fluoresces green when excited with ultraviolet or blue light without the addition of substrates or cofactors3,4,5. The gene for GFP has been cloned6 and is widely used in cellular biology7,8,9. In addition, the GFP gene has undergone modification for increased expression in plants, altered spectral properties, and improved fluorescence10,11. The mGFP4 variant features the removal of a plant-recognized cryptic intron10. Likewise, mGFP5er has been modified for cryptic intron mis-splicing and codon usage. It also contains intracellular targeting sequences10 and three amino acid substitutions. The mGFP5er gene is targeted to the endoplasmic reticulum by the fusion of a His-Asp-Glu-Leu polypeptide sequence to the C terminus and the fusion of Arabidopsis basic chitinase sequence to the N terminus in order to decrease hypothesized cytotoxicity of GFP10. Two mutations, V163A and S175G, enhance the folding of mGFP5er, especially at higher temperatures12, and an I167T substitution changes the ultraviolet (UV; 395 nm) and blue light (473 nm) maxima of mGFP5er to equal amplitudes. The sGFP gene is a synthetic human codon-optimized GFP gene with an S65T mutation in the chromophore, which leads to altered spectral qualities13,14. The S65T mutation and the optimization of codon usage yields about 100 times more fluorescence than wild-type GFP when excited by blue light13. The sGFP protein has a single excitation maximum of 490 nm and the emission maximum is 511 nm (ref. 4).
The green fluorescence produced by GFP is visually distinguishable from wild-type plants that autofluoresce reddish purple when excited by UV or blue light15, making it potentially useful for monitoring transgenes for agricultural and ecological applications16. However, for these purposes, GFP must be shown to fluoresce strongly in the field, have no fitness cost for synthesis, and be a functional indicator of the presence and expression of a second gene. In this study we have investigated these qualities by determining the effectiveness of monitoring the presence and expression of a Bacillus thuringiensis insecticidal crystal endotoxin-coding gene (Bt cry1Ac) in tobacco (Nicotiana tabacum) and oilseed rape (Brassica napus) using GFP fluorescence. This was acomplished by co-introducing into plants a plasmid containing the Bt cry1Ac and GFP genes. We found that in these transgenic plants, GFP fluorescence predicts Bt cry1Ac presence and expression. We also determined the costs and spectral qualities of GFP transgenic plants grown in the field.
Results
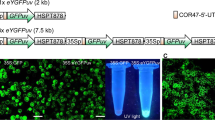
We recovered 150 mGFP5er-Bt cry1Ac transgenic tobacco and 20 transgenic oilseed rape regenerants. GFP-Bt segregation experiments were conducted on 14 independent T0 lines of GFP-Bt transgenic tobacco and oilseed rape to determine whether GFP segregation would match Bt segregation in T1 progeny. Following initial transformation of GFP-Bt into tobacco and oilseed rape, T1 progeny were germinated on soil and seedlings were screened under UV light for fluorescence. Mendelian segregation (3:1 ratio) was observed as a result of a single copy of the mGFP5er and Bt transgenes and high-level green fluorescence from GFP (Fig. 1). Bioassays confirmed that these lines killed corn earworm (CEW, Helicoverpa zea).
Transgenic (T1 progeny) containing Bt cry1Ac and GFP are shown growing on soil. (A) Over 300 T1 random tobacco plants and (B) 50 T1 oilseed rape plants were scored for GFP status by simple observation under UV light. Chi-squared analysis demonstrates that GFP and Bt are linked and are segregating in a 3:1 ratio (single copy of each of the transgenes). (C) Field-grown adult mGFP5-er transgenic tobacco are shown 90 days posttransplant. The transgenic plants are green and the null segregants are purple when illuminated by UV light.
Insect bioassays were conducted on leaf disks taken from multiple lines of GFP-Bt, mGFP5er, and wild-type tobacco and oilseed rape, representing a range of transgene expressors. Visual GFP fluorescence of GFP-Bt plants was negatively correlated with leaf defoliation by CEW in both tobacco and oilseed rape leaf samples. The GFP-Bt transgenic plant lines that were highly fluorescent were protected from insect herbivory by CEW. Plants with generally intermediate- or low-level green fluorescence suffered CEW damage to a significantly greater degree (Fig. 2). The B. rapa–oilseed rape hybrid retained both Bt cry1Ac and mGFPer genes. Furthermore, both genes were expressed to an equivalent extent in the hybrid and the oilseed rape parent (Fig. 3). Tobacco leaf disks that were defoliated by <10% also had significantly higher (p < 0.05) fluorescence spectrophotometry readings than those with greater defoliation (Fig. 4). Simple linear regression was significant (p = 0.03) but with a relatively low R2 (0.12). There were a few outliers in the data set. When the data set was truncated (discarding the points at >40% defoliation), statistical significance increased and was modeled well using polynomial regression (p = 0.0015, R2 = 0.46). The data demonstrate that selecting threshold fluorescence (say, 100,000 counts/s [c.p.s.]) would guarantee complete protection from CEW.
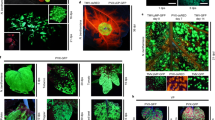
(A and B) Leaf disks of GFP-Bt tobacco and (C and D) GFP-Bt oilseed rape are shown. All leaf disks were subjected to corn earworm larvae (CEW, H. zea). (A and C) Leaf disks shown under white light. (B and D) Leaf disks photographed under UV light. (A and B) The genes in tobacco plants are: 1, mGFPer; 2, nontransgenic; 3, GFP-Bt. (C and D) Similar results are shown in oilseed rape: 1, nontransgenic oilseed rape (B. napus) ; 2, GFP-Bt oilseed rape; 3, Bt oilseed rape.
(A) 1: GFP-Bt B. napus parent leaf, 2: a B. rapa × B. napus hybrid leaf, and 3: a B. rapa leaf under white light. Each leaf was exposed to corn earworm larvae (CEW, H. zea). The B. napus parent and hybrid were strongly resistant to herbivory. In contrast, the B. rapa parent was susceptible to CEW damage. (B) The same three leaves illuminated by ultraviolet light. GFP fluorescence is detectable in both the B. napus and hybrid leaves but not in the B. rapa parent. The canola and hybrid plants expressed both the GFP and Bt genes at similar levels.
Leaf disks of a number of GFP-Bt transgenic plant lines with various expression of transgenes were subjected to CEW feeding. Significantly less GFP fluorescence was observed (P < 0 .05) in leaf disks that had high levels of defoliation (>10%). Means and standard deviations are shown. The inset shows the scatter plot of the raw data. a, b indicate significant differences at p = 0.05 (Tukey's HSD).
Protein synthesis in GFP-Bt plants used in bioassays and for spectrophotometry measurements was determined by protein blotting and immunostaining. The highest GFP synthesis level found in GFP-Bt transgenic tobacco was 0.2% of total extractable plant protein, whereas the highest Bt cry1Ac synthesis was 0.045%. In oilseed rape transgenics, maximal synthesis amounts of GFP and Bt were 0.25% and 0.075%, respectively. The GFP and Bt protein synthesis levels correlated with each other (P < 0.05, R = 0.86; n = 18).
Fluorescence and expression of GFP transgenics in the field.
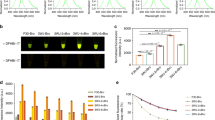
In a two-year field study (year 1: mGFP4, year 2: mGFP5er and sGFP), GFP fluorescence and possible GFP synthesis-related fitness drag were tested in transgenic tobacco for three GFP variants. Western blot analysis showed that >25% of mGFP4 transgenic plants produced at least 0.08% of total plant protein with a mean of 0.07% for high-fluorescing lines. The line with the highest expression of mGFP5er at midseason synthesized a mean of 0.14% GFP of total plant protein, whereas maximal sGFP production by a single line was a mean of 0.08%. High-fluorescent class lines of mGFP4, mGFP5er, and sGFP (except one line) synthesized significantly more protein than medium- and/or low-fluorescent classes (P < 0.05). However, there was a great deal of field-attributed variation observed in expression and fluorescence within transgenic events. All mGFP5er protein synthesis remained constant from midseason to season's end, and in four of the six sGFP lines, GFP production was lower at season's end.
After 90 days (midseason), mGFP4 transgenic plants were visually indistinguishable from wild-type tobacco plants under UV light; however, visual fluorescence of mGFP5er transgenic plants was easily detectable (Fig. 1C). The sGFP plants were also visually distinguishable from nontransgenic plants. Green fluorescent protein synthesis was correlated with fluorescence spectrophotometry measurements at midseason in all GFP transgenics (Fig. 5) but in only mGFP5er transgenics at the end of the growing season (p < 0.05, R2 = 0.12). There were differences in fluorescence spectrophotometry measurements of high, medium, and low classes and wild types in mGFP5er and sGFP transgenics at midseason and end of season but not in mGFP4 transgenics.
Regression lines of (A) year 1 mGFP4 (p = 0.4675, R2 = 0.0084) (B) year 2 mGFP5er (p = 0.001, R2 = 0.53), and (C) year 2 sGFP (p = 0.0001 R2 = 0.47) are represented. The mGFP5er and mGFP4 plants plus controls were excited at 385 nm (509 nm emission peaks recorded) and sGFP plants plus controls were excited at 490 nm (510 nm emission peaks recorded). High (▪●▴), medium (░), low (□○▵) events and controls (x) are shown.
Fitness of GFP transgenics.
Significantly different amounts of chlorophyll were synthesized between low- and medium-fluorescing classes of mGFP4 transgenics but not between the high-fluorescing class lines and controls. Chlorophyll amounts were not correlated with gene expression in mGFP4 plants (p = 0.98 R < 0.0001). Chlorophyll content was not significantly different from controls in mGFP5er (p = 0.86) and sGFP (p = 0.38) transgenics.
Control plants had higher biomass than mGFP4 transgenics; however, after the removal of an aberrant high-fluorescing event and its progeny, no difference was present in the analysis of variance. This event was characterized by low biomass and low seed production over a large range of actual GFP synthesis. We hypothesized that position effect of the transgene might explain the aberrant results. Synthesis of mGFP4 was not associated with biomass (p = 0.74, R2 = 0.002; Fig. 6). High-fluorescing class total seed weight in mGFP4 transgenics was also found to be significantly lower than that of medium-fluorescing, low-fluorescing, and wild-type classes; yet, seed weight was not associated with gene expression (p = 0.20, R2 = 0.22; Fig. 6).
Regression lines of (A) year 1 mGFP4 (B) year 2 mGFP5er, and (C) sGFP are represented. The mGFP5er and mGFP4 plants plus controls were excited at 385 nm (509 nm emission peaks recorded) and sGFP plants plus controls were excited at 490 nm (510 nm emission peaks recorded). High (▪●▴), medium (░), low (□○▵) events and controls (X) are shown. These classes were selected a priori before planting in the field.
One high-fluorescing sGFP line that expressed 0.06% GFP (the highest sGFP production level at the end of season) had significantly less biomass than all other plants. However, biomass was not associated with sGFP synthesis (p = 0.27, R2 = 0.015; Fig. 6). Total seed weight was different between sGFP plants and wild types (p = 0.04); yet, several high-expressing lines showed greater seed production than wild types, and sGFP synthesis was not related to seed weight (p = 0.10, R2 = 0.034; Fig. 6).
No significant differences were seen in average biomass (p = 0.12) or total seed weight of plants (p = 0.32) between mGFP5er transgenics and wild types. Chlorophyll content (p = 0.70, R = 0.02), biomass (p = 0.25, R2 = 0.022), and seed weight (p = 0.33, R2 = 0.016) were not associated with gene expression at both midseason (values given) and season's end (Fig. 6).
Discussion
There are several crops in the United States and Europe that have high rates of outcrossing between crop and wild relatives. For example, oilseed rape will hybridize with B. rapa and Raphanus raphanistrum under field conditions17,18,19. These hybrids readily backcross to the wild relative and may persist in the environment. However, this does not mean that transgenic free-living hosts will become "superweeds." Although Bt cry1Ac does enhance the fitness of oilseed rape in the field under nonagronomic conditions, it has not been demonstrated to increase weediness of oilseed rape20. It is not known whether relatives of transgenic oilseed rape will cause ecological shifts in unmanaged ecosystems (i.e., nature). Until agriculturally relevant and realistically sized field experiments are performed, the risk of transgenic fitness-enhanced oilseed rape may only be conjectured. A second example of a potentially risky commercial release is fitness-enhanced transgenic sunflower (Helianthus annuus)21. The center of diversity for sunflower is the midwestern United States, and there is concern—though not experimentally validated—that release of a gene that can be acted upon by natural selection would decrease the genetic diversity of wild sunflower (same species).
In both of these examples, experimental and subsequent commercial monitoring of certain genes is needed. This study shows that GFP would be useful for such a purpose, as GFP linked to a second transgene can indicate the presence of the second gene, and that both transgenes are expressed in crop progeny and a crop–wild relative hybrid. Moreover, it potentially could be used at the scale of large field experiments for commercial agricultural releases. Although illuminating plants with UV light at night is awkward, it can provide a rapid and accurate screen. Alternatively, a simple, portable single-wavelength fluorescence spectrophotometer could be used even in daylight.
In this study, insect antibiosis mediated by Bt Cry1Ac could be predicted by green fluorescence from GFP. In this case, CEW is sensitive to Bt Cry1Ac toxin. For example, in transgenic soybean 0.005% Bt Cry1Ac synthesis limited CEW defoliation to approximately 1% (ref. 22). A priori, we would expect a more linear relationship when the target insect is less susceptible to the transgenic toxin. We envisage GFP fluorescence to be most useful as a threshold marker [i.e., presence (+)/absence (–)].
Existing technologies for monitoring transgene expression, such as immunological or histological methods, are expensive and require tissue sampling. There are several situations in which a grower or researcher may be interested in assessing the expression of a second transgene in a large number of samples. For example, a grower may be concerned about adequate expression of an herbicide tolerance gene before herbicide treatment. Another example is the maintenance of a Bt-resistance management strategy in which a high dose of Bt is required. If a GFP monitoring system is to function in the field, high-level expression of transgenes is necessary.
There has been anecdotal evidence reported that GFP is cytotoxic10, which would make it impractical for experimental or commercial releases. We have shown that when GFP is synthesized and resides in the cytoplasm it does not accumulate or fluoresce as well as an endoplasmic reticulum–targeted GFP. However, none of the GFPs tested were toxic to tobacco plants growing in the field.
As presented, there are potential limitations to a GFP monitoring system. For example, the transgenes might become unlinked over multiple generations. Another limitation is that differences in relative expression of the two transgenes can occur. However, if the genes are integrated in a transcriptionally active area of the genome, a priori, they will both be expressed to a high degree23. GFP in transcriptional or translational fusions would address these potential limitations.
Experimental protocol
Plant expression vectors. All vectors contained GFP genes under the control of the cauliflower mosaic virus 35S promoter and an NOS (nopaline synthase) terminator. All plasmids used an npt II cassette for selection in plants under the control of the nos terminator. The mGFP4 gene was the kind gift of Jim Haseloff in the pBin19 35S-mGFP4 plasmid10. The mGFP5er gene in the pBin-m-GFP5-er plasmidwas also a gift of Jim Haseloff 10,12. The sGFP gene in the Bin19-35S-SGFP-TYG-nos plasmid was the kind gift of Jen Sheen by way of C.S. Prakash13.
The mGFP5er-Bt cry1Ac vector (pSAM12) was constructed as follows: The mGFP4 cassette from the pBin19 35S-mGFP4 plasmid was excised by an EcoRI and HindIII digestion and ligated between two tobacco RB7 scaffold attachment regions (SARs)23 in a pBluescript II KS (Stratagene, La Jolla, CA) vector. All bands used to make constructs were gel purified using the GENECLEAN kit (Bio 101, Carlsbad, CA). Next, the Bt cry1Ac gene (synthetic, codon-optimized, truncated) with a 35S promoter and an orf25 terminator was liberated from pSG/BT (ref. 22) by BglII digestion and ligated between the SAR regions downstream of the mGFP4 cassette into a BamHI site. In the pBin-m-GFP5-er plasmid, mGFP5er was digested with SacI, linearizing the vector. The mGFP4/Bt cry1Ac construct was partially digested with SacI with the band containing the nos terminator from mGFP4 and the entire Bt cry1Ac followed by a single SAR region. This fragment was ligated into the linearized mGFP5-ER fragment, and the final construct was named pSAM12, which contained npt II, mGFP5er, and Bt cry1Ac cassettes. These cassettes were oriented in head-tail fashion.
Plant transformation and hybridization.
pSAM12 and three other expression vectors containing either the mGFP4, mGFP5er, or sGFP genes were transformed into tobacco (Nicotiana tabacum cv. Xanthi)24 and oilseed rape (B. napus cv Westar)25 using Agrobacterium tumefaciens (strain GV3850) mediated transformation. Putative transgenics were then screened by using a handheld UV light (UVP model B-100AP 100 W: 365 nm) or under a fluorescent microscope (485 nm) to assess green fluorescence. In some experiments, transgenics were classified as high-, medium-, low-, or nonfluorescing. Oilseed rape GFP-Bt transgenics were hybridized with B. rapa (wild accessions) from Montana or California (the kind gifts of Randy Linder and Art Weiss, respectively) by hand crosses. The putative hybrid seeds harvested from B. rapa were germinated and screened by visual observation under the illumination of a UV light.
Insect bioassays.
Two leaf disks (2 cm diameter) from the second and fourth leaves of the apical meristem from 31 independent GFP-Bt transgenic tobacco plants were placed in glass petri dishes on moist filter paper. Ten 4-h-old neonate larvae of corn earworm (H. zea) were placed on each disk for one week, at which time the amount of leaf disk defoliation and the number of alive and dead insects were quantified. Eighteen T1 plants containing seven null segregant lines plus 13 different tobacco T0 lines were used in addition to three mGFP5er transgenic tobacco lines that served as negative controls. A similar experiment was performed using a total of 20 GFP oilseed rape, GFP-Bt oilseed rape, B. rapa, and GFP-Bt B. rapa × oilseed rape lines containing two replicate leaf samples.
Fluorescence detection.
Quantification of GFP fluorescence was achieved using a Fluoromax-2 fluorescence spectrophotometer (Instruments S.A., Edison, NJ) using DataMax and GRAMS/386 software (Galactic Industries Corporation, Salem, NH). A 2 m randomized, bifurcated fiberoptic cable was used in vivo to transmit excitation light and detect emission transmission from intact leaves. Fluorescence spectrometry was performed on all leaves in insect bioassays preceding insect addition. Fluorescence spectrophotometry was performed in the middle and the end (mGFP5er and sGFP study only) of the growing season on the third leaf from the apex of plants in the field study. Transgenic plants containing the mGFP4 and mGFP5er genes were excited at 385 nm, and emission was detected at 509 nm. The sGFP transgenic plants were excited at 490 nm and measured at 510 nm. Wild-type plants were excited and measured at both sets of wavelengths. Wild-type, mGFP4, and mGFP5er transgenic plants in the field were also visually assayed in the field at night by using a portable handheld UV light (UVP B-model 100AP; 100 W; 365 nm, UVP, Upland, CA). Plants were classified as high-, medium-, low-, or nonfluorescing in the field.
Protein analysis.
Total protein was extracted and analyzed using western blots from GFP transgenic16 and GFP- Bt cry1Ac transgenic plants23 using Bt- and GFP-specific polyclonal antibodies.
Field designs.
In the first season of study, 240 transgenic plants of various fluorescing classes and 80 wild-type tobacco plants were placed in the field using a Latin square design26. The design consisted of 69 plants from 8 T1 and 171 plants from 16 T2 plant lines. These were planted in eight rows with 40 plants per row. Each row consisted of four plots (high-, medium-, and low-fluorescing and wild-type plants) with 10 plants each. The fluorescent classes were determined by apparent fluorescence of the primary transformant. In the second year 378 sGFP and 378 mGFP5er T2 plants were placed in the field with 126 wild types for another growing season using a similar Latin square design. One field consisted of only mGFP5er transgenics and controls, another of sGFP transgenics and controls, and a center field consisted of both transgenic types and controls. Each field consisted of seven pairs of rows consisting of 42 individual plants in a series of 2 × 3 plant plots (six from each transgenic line and wild types). Wild-type tobacco (cv. Xanthi) plants were grown at the end of each row, and full rows of control plants were also grown before the first and after the last row in all three fields to serve as environmental buffers. Plants were grown under field conditions from early June to mid-October at the Upper Piedmont Research Station (Reidsville, NC).
Chlorophyll content and reproductive/vegetative output.
Relative chlorophyll concentrations were estimated in plants using a Minolta SPAD-502 Chlorophyll Meter (Spectrum Technologies, Plainfield, IL). Total seed weight per plant (mGFP4 experiment) after the first year was estimated by counting the total number of seed capsules in all plants at the end of the season and removing all capsules from one plant per plot. Seeds were collected from 20 random capsules and weighed. This average total seed weight per capsule was then multiplied by the total number of capsules for each plant. In the second year of study (sGFP and GFP5er experiment) total seed weights were measured after the growing season for each plant. Capsules were taken from the field and allowed to dry for seven days before measuring in both cases. At the end of both growing seasons total wet plant biomass data were taken for all plants by harvesting the plants at ground level and then weighing the plant while in the field.
References
Raybould, A.F. & Gray, A.J. Genetically modified crops and hybridization with wild relatives: a UK perspective. J. Appl. Ecol. 30, 199–219 (1993).
Rogers, H.J. & Parkes, H.C. Transgenic plants and the environment. J.Exp.Bot. 46, 467–488 (1995).
Stewart, C.N., Jr. Monitoring transgenic plants using in vivo markers. Nat. Biotechnol. 14, 682 (1996).
Heim, R. & Tsien, R.Y. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr. Biol. 6, 178–182 (1996).
Yang, F., Moss, L.G. & Phillips, G.N., Jr. The molecular structure of green fluorescent protein. Nat. Biotechnol. 14, 1246–1251 (1996).
Prasher, D.C., Eckenrode, V.K., Ward, W.W., Prendergast, F.G. & Cormier, M.J. Primary structure of the Aequorea victoria green fluorescent protein. Gene 111, 229–233 (1992).
Grebenok, R.J. et al. Green-fluorescent protein fusions for efficient characterization of nuclear targeting. Plant J. 11, 573–586 (1997).
Kohler, R.H., Zipfel, W.R., Webb, W.W. & Hanson, M.R. The green fluorescent protein as a marker to visualize plant mitochondria in vivo. Plant J. 11, 613–621 (1997).
Plautz, J.D. et al. Green fluorescent protein and its derivatives as versatile markers for gene expression in living Drosophila melanogaster, plant and mammalian cells. Gene 173, 83–87 (1996).
Haseloff, J., Siemering, K.R., Prasher, D.C. & Hodge, S. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94, 2122–2127 (1997).
Reichel, C. et al. Enhanced green fluorescence by the expression of an Aequorea victoria green fluorescent protein mutant in mono- and dicotyledonous plant cells. Proc. Natl. Acad. Sci. USA 93, 5888–5893 (1996).
Siemering, K.R., Golbik, R., Sever, R. & Haseloff, J. Mutations that suppress the thermosensitivity of green fluorescent protein. Curr. Biol. 6, 1653–1663 (1996).
Chiu, W.L., Niwa, Y., Zeng, W., Hirano, T., Kobayashi, H. & Sheen, J. Engineered GFP as a vital reporter in plants. Curr. Biol. 6, 325–330 (1996).
Haas, J., Park, E.-C. & Seed, B. Codon usage limitation in the expression if HIV-1 envelope glycoprotein. Curr. Biol. 6, 315–324 (1996).
An, G. High efficiency transformation of cultured tobacco cells. Plant Physiol. 79, 568–570 (1985).
Leffel, S.M., Mabon, S.A. & Stewart, C.N., Jr. Applications of green fluorescent protein in plants. BioTechniques 23, 912–918 (1997).
Chevre, A.M., Eber, F., Baranger, A. & Renard, M. Gene flow from transgenic crops. Nature 389, 924–924 (1997).
Jørgensen, R.B. & Andersen, B. Spontaneous hybridization between oilseed rape (Brassica napus) and weedy B. campestris (Brassicaceae): a risk of growing genetically modified oilseed rape. Am. J. Bot. 81, 1620–1626 (1994).
Mikkelsen, T.R., Andersen, B. & Jørgensen, R.B. The risk of crop transgene spread. Nature 380, 31 (1996).
Stewart, C.N., Jr., All, J.N., Raymer, P.L. & Ramachandran, S. Increased fitness of transgenic insecticidal rapeseed under insect selection pressure. Mol. Ecol. 6, 773–779 (1997).
Whitton, J., Wolf, D.E., Arias, D.M., Snow, A.A. & Rieseberg, L.H. The persistence of cultivar alleles in wild populations of sunflowers five generations after hybridization. Theor. Appl. Genet. 95, 33–40 (1997).
Stewart, C.N., Jr. et al. Genetic transformation, recovery, and characterization of fertile soybean transgenic for a synthetic Bacillus thuringiensis cryIAc gene. Plant Physiol. 112, 121–129 (1996).
Allen, G.C. et al. High-level transgene expression in plant cells: effects of a strong scaffold attachment region from tobacco. Plant Cell 8, 899–913 (1996).
Horsch, R.B. et al. A simple and general method for transferring genes into plants. Science 227, 1229–1231 (1985).
Stewart, C.N., Jr. et al. Insect control and dosage effects in transgenic canola, Brassica napus L. (Brassicaceae) containing a synthetic Bacillus thuringiensis cryIAc gene. Plant Physiol. 112, 115–120 (1996).
Sokal, R.R. & Rohlf, F.J. in Biometry Vol. 2, 372–399 (W.H. Freeman and Company, New York; 1981).
Acknowledgements
We thank Jim Haseloff for the gift of the mGFP4 and mGFP5er constructs. We thank Jen Sheen and C.S. Prakash for the gift of the sGFP gene. We thank Guy Cardineau and Dow AgroSciences for the gift of the Bt cry1Ac gene, protein, and antisera. Research was funded by the USDA Biotechnology Risk Assessment grant 98-33522-6797, and NSF grant 9604528. We also thank Jimmy Meadows and Joe French at the Upper Piedmont Expression Research Station, as well as Sheila Branch, Tanya Chalk, Dean Chamberlain, Richard Emanuel, Mark Hens, Laura Hudson, Kevin Markham, Reggie Millwood, Alana Riddick, Shana Stovall, Elizabeth Tomlin, and Elyse Williams, for help in both the field and the lab.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Harper, B., Mabon, S., Leffel, S. et al. Green fluorescent protein as a marker for expression of a second gene in transgenic plants. Nat Biotechnol 17, 1125–1129 (1999). https://doi.org/10.1038/15114
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/15114
This article is cited by
-
Optimization and establishment of Agrobacterium-mediated transformation in alfalfa (Medicago sativa L.) using eGFP as a visual reporter
Plant Cell, Tissue and Organ Culture (PCTOC) (2024)
-
Generation of brilliant green fluorescent petunia plants by using a new and potent fluorescent protein transgene
Scientific Reports (2018)
-
Pentapeptide-insertion scanning mutational analysis of turkey herpesvirus HVT063 reveals residues important for its RNA silencing suppression activity
Archives of Virology (2018)
-
Evaluation of a MdMYB10/GFP43 fusion gene for its suitability to act as reporter gene in promoter studies in Fragaria vesca L. ‘Rügen’
Plant Cell, Tissue and Organ Culture (PCTOC) (2017)
-
An orange fluorescent protein tagging system for real-time pollen tracking
BMC Research Notes (2013)