Abstract
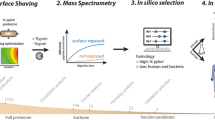
We describe a proteomic approach for identifying bacterial surface-exposed proteins quickly and reliably for their use as vaccine candidates. Whole cells are treated with proteases to selectively digest protruding proteins that are subsequently identified by mass spectrometry analysis of the released peptides. When applied to the sequenced M1_SF370 group A Streptococcus strain, 68 PSORT-predicted surface-associated proteins were identified, including most of the protective antigens described in the literature. The number of surface-exposed proteins varied from strain to strain, most likely as a consequence of different capsule content. The surface-exposed proteins of the highly virulent M23_DSM2071 strain included 17 proteins, 15 in common with M1_SF370. When 14 of the 17 proteins were expressed in E. coli and tested in the mouse for their capacity to confer protection against a lethal dose of M23_DSM2071, one new protective antigen (Spy0416) was identified. This strategy overcomes the difficulties so far encountered in surface protein characterization and has great potential in vaccine discovery.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Lindahl, G., Stalhammar-Carlemalm, M. & Areschoug, T. Surface proteins of Streptococcus agalactiae and related proteins in other bacterial pathogens. Clin. Microbiol. Rev. 18, 102–127 (2005).
Courtney, H.S., Dale, J.B. & Hasty, D.L. in Handbook of Bacterial Adhesion: Principles, Methods and Applications (eds. An, Y.N. & Friedman, R.J.) 553–579 (Human Press, Totowa, NJ, 2002).
Lin, J., Huang, S. & Zhang, Q. Outer membrane proteins: key players for bacterial adaptation in host niches. Microbes Infect. 4, 325–331 (2002).
Niemann, H.H., Schubert, W.D. & Heinz, D.W. Adhesins and invasins of pathogenic bacteria: a structural view. Microbes Infect. 6, 101–112 (2004).
Ton-That, H., Marraffini, L.A. & Schneewind, O. Protein sorting to the cell wall envelope of Gram-positive bacteria. Biochim. Biophys. Acta 1694, 269–278 (2004).
Janulczyk, R. & Rasmussen, M. Improved pattern for genome-based screening identifies novel cell wall–attached proteins in gram-positive bacteria. Infect. Immun. 69, 4019–4026 (2001).
Galperin, M.Y. & Koonin, E.V. Searching for drug targets in microbial genomes. Curr. Opin. Biotechnol. 10, 571–578 (1999).
Pizza, M. et al. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science 287, 1816–1820 (2000).
Maione, D. et al. Identification of a universal group B Streptococcus vaccine by multiple genome screen. Science 309, 148–150 (2005).
Nakai, K. Protein sorting signals and prediction of subcellular localization. Adv. Protein Chem. 54, 277–344 (2000).
Doytchinova, I.A., Taylor, P. & Flower, D.R. Proteomics in vaccinology and immunobiology: an informatics perspective of the immunone. J. Biomed. Biotechnol. 2003, 267–290 (2003).
Molloy, M.P. et al. Proteomic analysis of the Escherichia coli outer membrane. Eur. J. Biochem. 267, 2871–2881 (2000).
Phadke, N.D. et al. Analysis of the outer membrane proteome of Caulobacter crescentus by two-dimensional electrophoresis and mass spectrometry. Proteomics 1, 705–720 (2001).
Molloy, M.P. et al. Profiling the alkaline membrane proteome of Caulobacter crescentus with two-dimensional electrophoresis and mass spectrometry. Proteomics 2, 899–910 (2002).
Nouwens, A.S. et al. Complementing genomics with proteomics: the membrane subproteome of Pseudomonas aeruginosa PAO1. Electrophoresis 21, 3797–3809 (2000).
Rhomberg, T.A. et al. Proteomic analysis of the sarcosine-insoluble outer membrane fraction of the bacterial pathogen Bartonella henselae. Proteomics 4, 3021–3033 (2004).
Sabarth, N. et al. Identification of surface proteins of Helicobacter pylori by selective biotinylation, affinity purification, and two-dimensional gel electrophoresis. J. Biol. Chem. 277, 27896–27902 (2002).
Guina, T. et al. Proteomic analysis of Pseudomonas aeruginosa grown under magnesium limitation. J. Am. Soc. Mass Spectrom. 14, 742–751 (2003).
Ferretti, J.J. et al. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl Acad. Sci. USA 98, 4658–4663 (2001).
Marques, M.A., Chitale, S., Brennan, P.J. & Pessolani, M.C. Mapping and identification of the major cell wall-associated components of Mycobacterium leprae. Infect. Immun. 66, 2625–2631 (1998).
Dallo, S.F., Kannan, T.R., Blaylock, M.W. & Baseman, J.B. Elongation factor Tu and E1 beta subunit of pyruvate dehydrogenase complex act as fibronectin binding proteins in Mycoplasma pneumoniae. Mol. Microbiol. 46, 1041–1051 (2002).
Spence, J.M. & Clark, V.L. Role of ribosomal protein L12 in gonococcal invasion of Hec1B cells. Infect. Immun. 68, 5002–5010 (2000).
Kurar, E. & Splitter, G.A. Nucleic acid vaccination of Brucella abortus ribosomal L7/L12 gene elicits immune response. Vaccine 15, 1851–1857 (1997).
Tomoyasu, T. et al. Topology and subcellular localization of FtsH protein in Escherichia coli. J. Bacteriol. 175, 1352–1357 (1993).
Akiyama, Y., Kihara, A., Mori, H., Ogura, T. & Ito, K. Roles of the periplasmic domain of Escherichia coli FtsH (HflB) in protein interactions and activity modulation. J. Biol. Chem. 273, 22326–22333 (1998).
Amara, R.R., Shanti, S. & Satchidanandam, V. Characterization of novel immunodominant antigens of Mycobacterium tuberculosis. Microbiology 144, 1197–1203 (1998).
Padmalayam, I., Anderson, B., Kron, M., Kelly, T. & Baumstark, B. The 75-kilodalton antigen of Bartonella bacilliformis is a structural homolog of the cell division protein FtsZ. J. Bacteriol. 179, 4545–4552 (1997).
Paterson, G.K. & Mitchell, T.J. The biology of Gram-positive sortase enzymes. Trends Microbiol. 12, 89–95 (2004).
Lancefield, R.C. Studies on the antigenic composition of group A hemolytic Streptococci. J. Exp. Med. 78, 465–476 (1943).
Mora, M. et al. Group A Streptococcus produce pilus-like structures containing protective antigens and Lancefield T antigens. Proc. Natl Acad. Sci. USA 102, 15641–15646 (2005).
Stalhammar-Carlemalm, M., Areschoug, T., Larsson, C. & Lindahl, G. The R28 protein of Streptococcus pyogenes is related to several group B streptococcal surface proteins, confers protective immunity and promotes binding to human epithelial cells. Mol. Microbiol. 33, 208–219 (1999).
Reid, S.D. et al. Postgenomic analysis of four novel antigens of group A Streptococcus: growth phase-dependent gene transcription and human serologic response. J. Bacteriol. 184, 6316–6324 (2002).
McMillan, D.J. et al. Identification and assessment of new vaccine candidates for group A streptococcal infections. Vaccine 22, 2783–2790 (2004).
Ji, Y., Carlson, B., Kondagunta, A. & Cleary, P.P. Intranasal immunization with C5a peptidase prevents nasopharyngeal colonization of mice by the group A Streptococcus. Infect. Immun. 65, 2080–2087 (1997).
Massell, B.F., Michael, J.G., Amezcua, J. & Siner, M. Secondary and apparent primary antibody responses after group A streptococcal vaccination of 21 children. Appl. Microbiol. 16, 509–518 (1968).
Dale, J.B., Chiang, E.Y., Liu, S., Courtney, H.S. & Hasty, D.L. New protective antigen of group A Streptococci. J. Clin. Invest. 103, 1261–1268 (1999).
Kawabata, S. et al. Systemic and mucosal immunizations with fibronectin-binding protein FBP54 induce protective immune responses against Streptococcus pyogenes challenge in mice. Infect. Immun. 69, 924–930 (2001).
Guzman, C.A., Talay, S.R., Molinari, G., Medina, E. & Chhatwal, G.S. Protective immune response against Streptococcus pyogenes in mice after intranasal vaccination with the fibronectin-binding protein SfbI. J. Infect. Dis. 179, 901–906 (1999).
Kuo, C.F. et al. Role of streptococcal pyrogenic exotoxin B in the mouse model of group A streptococcal infection. Infect. Immun. 66, 3931–3935 (1998).
Roggiani, M. et al. Toxoids of streptococcal pyrogenic exotoxin A are protective in rabbit models of streptococcal toxic shock syndrome. Infect. Immun. 68, 5011–5017 (2000).
Lei, B., Liu, M., Chesney, G.L. & Musser, J.M. Identification of new candidate vaccine antigens made by Streptococcus pyogenes: purification and characterization of 16 putative extracellular lipoproteins. J. Infect. Dis. 189, 79–89 (2004).
Cunningham, M.W. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13, 470–511 (2000).
Cole, J.N. et al. Surface analyses and immune reactivities of major cell wall–associated proteins of group A Streptococcus. Infect. Immun. 73, 3137–3146 (2005).
Lancefield, R.C. Persistence of type-specific antibodies in man following infection with group A streptococci. J. Exp. Med. 110, 271–292 (1959).
Lancefield, R.C. Current knowledge of type-specific M antigens of group A Streptococci. J. Immunol. 89, 307–313 (1962).
Hu, M.C. et al. Immunogenicity of a 26-valent group A streptococcal vaccine. Infect. Immun. 70, 2171–2177 (2002).
Fernandez-Espla, M.D., Garault, P., Monnet, V. & Rul, F. Streptococcus thermophilus cell wall–anchored proteinase: release, purification, and biochemical and genetic characterization. Appl. Environ. Microbiol. 66, 4772–4778 (2000).
Harris, T.O., Shelver, D.W., Bohnsack, J.F. & Rubens, C.E. A novel streptococcal surface protease promotes virulence, resistance to opsonophagocytosis, and cleavage of human fibrinogen. J. Clin. Invest. 111, 61–70 (2003).
Lei, B., Mackie, S., Lukomski, S. & Musser, J.M. Identification and immunogenicity of group A Streptococcus culture supernatant proteins. Infect. Immun. 68, 6807–6818 (2000).
Acknowledgements
We thank J.M. Musser and G. Orefici for providing some of the strains used in this work and Marco Tortoli for his expert assistance in animal studies. We are also grateful to Giorgio Corsi for the artwork and Antonietta Maiorino for her expert secretarial assistance. Manuel J. Rodríguez-Ortega had an Intra-European Marie Curie postdoctoral fellowship from the EU. This work was in part supported by National Institutes of Health/ National Institute of Allergy and Infectious Diseases grant number 5U1060595-02, and in part by the Italian Ministry of Education, University and Research grant no. 10811.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
All authors work in a private company as either full-time employees or contract scientists and, in particular, G.G. and J.L.T. are holders of company stock options.
Supplementary information
Supplementary Table 1
Surfome of Group A Streptococcus M1_SF370 strain. (PDF 45 kb)
Supplementary Table 2
List of M1_SF370 surfome proteins grouped in degree-of-confidence categories. (PDF 14 kb)
Supplementary Table 3
Surfome of Group A Streptococcus M3_CDCSS90 strain and comparison with the M1_SF370 surfome. (PDF 13 kb)
Rights and permissions
About this article
Cite this article
Rodríguez-Ortega, M., Norais, N., Bensi, G. et al. Characterization and identification of vaccine candidate proteins through analysis of the group A Streptococcus surface proteome. Nat Biotechnol 24, 191–197 (2006). https://doi.org/10.1038/nbt1179
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt1179
This article is cited by
-
Structure–activity studies of Streptococcus pyogenes enzyme SpyCEP reveal high affinity for CXCL8 in the SpyCEP C-terminal
Scientific Reports (2023)
-
Pathoproteomik des humanpathogenen Pilzes Aspergillus fumigatus
BIOspektrum (2023)
-
Identification of surface proteins in a clinical Staphylococcus haemolyticus isolate by bacterial surface shaving
BMC Microbiology (2020)
-
Cross-serotype protection against group A Streptococcal infections induced by immunization with SPy_2191
Nature Communications (2020)
-
Predicting Promiscuous T Cell Epitopes for Designing a Vaccine Against Streptococcus pyogenes
Applied Biochemistry and Biotechnology (2019)