Abstract
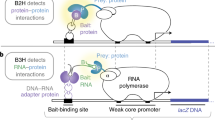
The DNA-binding specificities of transcription factors can be used to computationally predict cis-regulatory modules (CRMs) that regulate gene expression1. However, the absence of specificity data for the majority of transcription factors limits the widespread implementation of this approach. We have developed a bacterial one-hybrid system that provides a simple and rapid method to determine the DNA-binding specificity of a transcription factor. Using this technology, we successfully determined the DNA-binding specificity of seven previously characterized transcription factors and one novel transcription factor, the Drosophila melanogaster factor Odd-skipped. Regulatory targets of Odd-skipped were successfully predicted using this information, demonstrating that the data produced by the bacterial one-hybrid system are relevant to in vivo function.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Berman, B.P. et al. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl. Acad. Sci. USA 99, 757–762 (2002).
Roulet, E. et al. High-throughput SELEX–SAGE method for quantitative modeling of transcription-factor binding sites. Nat. Biotechnol. 20, 831–835 (2002).
Bulyk, M.L., Huang, X., Choo, Y. & Church, G.M. Exploring the DNA-binding specificities of zinc fingers with DNA microarrays. Proc. Natl. Acad. Sci. USA 98, 7158–7163 (2001).
Mukherjee, S. et al. Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nat. Genet. 36, 1331–1339 (2004).
Lee, T.I. et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298, 799–804 (2002).
Liu, X., Noll, D.M., Lieb, J.D. & Clarke, N.D. DIP-chip: rapid and accurate determination of DNA-binding specificity. Genome Res. 15, 421–427 (2005).
Wilson, T.E., Fahrner, T.J., Johnston, M. & Milbrandt, J. Identification of the DNA binding site for NGFI-B by genetic selection in yeast. Science 252, 1296–1300 (1991).
Deplancke, B., Dupuy, D., Vidal, M. & Walhout, A.J. A gateway-compatible yeast one-hybrid system. Genome Res. 14, 2093–2101 (2004).
Joung, J.K., Ramm, E.I. & Pabo, C.O. A bacterial two-hybrid selection system for studying protein-DNA and protein-protein interactions. Proc. Natl. Acad. Sci. USA 97, 7382–7387 (2000).
Dove, S.L., Joung, J.K. & Hochschild, A. Activation of prokaryotic transcription through arbitrary protein-protein contacts. Nature 386, 627–630 (1997).
Bailey, T.L. & Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36 (1994).
Wolfe, S.A., Greisman, H.A., Ramm, E.I. & Pabo, C.O. Analysis of zinc fingers optimized via phage display: evaluating the utility of a recognition code. J. Mol. Biol. 285, 1917–1934 (1999).
Voz, M.L., Agten, N.S., Van de Ven, W.J. & Kas, K. PLAG1, the main translocation target in pleomorphic adenoma of the salivary glands, is a positive regulator of IGF-II. Cancer Res. 60, 106–113 (2000).
Walhout, A.J. & Vidal, M. A genetic strategy to eliminate self-activator baits prior to high-throughput yeast two-hybrid screens. Genome Res. 9, 1128–1134 (1999).
Greisman, H.A. & Pabo, C.O. A general strategy for selecting high-affinity zinc finger proteins for diverse DNA target sites. Science 275, 657–661 (1997).
Senger, K. et al. Immunity regulatory DNAs share common organizational features in Drosophila. Mol. Cell 13, 19–32 (2004).
Kovall, R.A. & Hendrickson, W.A. Crystal structure of the nuclear effector of Notch signaling, CSL, bound to DNA. EMBO J. 23, 3441–3451 (2004).
Tun, T. et al. Recognition sequence of a highly conserved DNA binding protein RBP-J kappa. Nucleic Acids Res. 22, 965–971 (1994).
Jun, S. & Desplan, C. Cooperative interactions between paired domain and homeodomain. Development 122, 2639–2650 (1996).
Melnikova, I.N., Crute, B.E., Wang, S. & Speck, N.A. Sequence specificity of the core-binding factor. J. Virol. 67, 2408–2411 (1993).
Golling, G., Li, L., Pepling, M., Stebbins, M. & Gergen, J.P. Drosophila homologs of the proto-oncogene product PEBP2/CBF beta regulate the DNA-binding properties of Runt. Mol. Cell. Biol. 16, 932–942 (1996).
Sosinsky, A., Bonin, C.P., Mann, R.S. & Honig, B. Target Explorer: an automated tool for the identification of new target genes for a specified set of transcription factors. Nucleic Acids Res. 31, 3589–3592 (2003).
Riddihough, G. & Ish-Horowicz, D. Individual stripe regulatory elements in the Drosophila hairy promoter respond to maternal, gap, and pair-rule genes. Genes Dev. 5, 840–854 (1991).
Saulier-Le Drean, B., Nasiadka, A., Dong, J. & Krause, H.M. Dynamic changes in the functions of Odd-skipped during early Drosophila embryogenesis. Development 125, 4851–4861 (1998).
Serebriiskii, I. & Joung, J. in Protein-Protein Interactions: A Molecular Cloning Manual (ed. E. Golemis) 93–142, (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 2001).
Lutz, R. & Bujard, H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1–I2 regulatory elements. Nucleic Acids Res. 25, 1203–1210 (1997).
Liu, X., Brutlag, D.L. & Liu, J.S. BioProspector: discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac. Symp. Biocomput. 6, 127–138 (2001).
Schneider, T.D. & Stephens, R.M. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18, 6097–6100 (1990).
Crooks, G.E., Hon, G., Chandonia, J.M. & Brenner, S.E. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004).
Tautz, D. & Pfeifle, C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma 98, 81–85 (1989).
Acknowledgements
We would like to thank Keith Joung, Jessica Hurt, Carl Pabo and Hermann Bujard for precursor plasmids and strains. Henry Krause for providing the HSodd2 flies. Lucio Castilla, Sean Landrette, Marian Walhout, Marc Freeman, Tony Ip and the Drosophila Genomic Resource Center for various cDNAs. Nadine McGinnis and Robin Smith for technical support. Marian Walhout and Keith Joung for very helpful discussions. We thank the UCSC genome bioinformatics site and the Institute for Genomic Research, the Genome Sequencing Center at Washington University, Agencourt Bioscience Corporation and HGSC at Baylor College of Medicine for access to and analysis of unpublished Drosophila genome data. S.A.W. and X.M. were supported in part by the Concern Foundation and National Institutes of Health (NIH) grant 1R01GM068110; M.H.B. was supported in part by a Basil O'Connor Starter Research Award from the March of Dimes Birth Defects Foundation and by the American Cancer Society grant RSG-05-026-01-CCG. This work was supported by NIH grant 1R01GM068110 (S.A.W.). This work was supported by NIH grant 1R01GM068110 (S.A.W.) and ACS grant RSG-05-026-01-CCG (M.H.B.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors have a pending patent application on related subject matter.
Supplementary information
Supplementary Fig. 1
Comparison of the number of self-activating clones in the original and purified library. (PDF 418 kb)
Supplementary Fig. 2
The Runt/Bgb heterodimer is required for reporter activation. (PDF 450 kb)
Supplementary Fig. 3
Growth rates of cells containing different Odd bait-prey combinations. (PDF 754 kb)
Supplementary Fig. 4
Schematic representation of the conserved Odd binding sites near hairy, paired, gooseberry and Goosecoid. (PDF 153 kb)
Supplementary Table 1
Details of the selections performed with each bait using the B1H system. (PDF 20 kb)
Supplementary Table 2
Unique insert sequences from the prey isolated using the B1H system with each bait. (PDF 61 kb)
Supplementary Table 3
Search results using a PWM for Odd built in Target Explorer. (PDF 82 kb)
Rights and permissions
About this article
Cite this article
Meng, X., Brodsky, M. & Wolfe, S. A bacterial one-hybrid system for determining the DNA-binding specificity of transcription factors. Nat Biotechnol 23, 988–994 (2005). https://doi.org/10.1038/nbt1120
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt1120
This article is cited by
-
EGR1-CCL2 Feedback Loop Maintains Epithelial-Mesenchymal Transition of Cisplatin-Resistant Gastric Cancer Cells and Promotes Tumor Angiogenesis
Digestive Diseases and Sciences (2022)
-
Combining transposon mutagenesis and reporter genes to identify novel regulators of the topA promoter in Streptomyces
Microbial Cell Factories (2021)
-
Engineered dual selection for directed evolution of SpCas9 PAM specificity
Nature Communications (2021)
-
An improved bind-n-seq strategy to determine protein-DNA interactions validated using the bacterial transcriptional regulator YipR
BMC Microbiology (2020)
-
A Hox-TALE regulatory circuit for neural crest patterning is conserved across vertebrates
Nature Communications (2019)