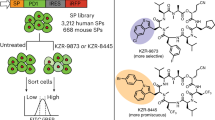
Abstract
There is great interest in the identification of synthetic molecules that are capable of manipulating protein-protein interactions in living cells. Peptides, unlike other classes of small molecules, have binding properties appropriate for this application, but most are poorly cell permeable and sensitive to proteases. Therefore, considerable effort has been expended in the development of libraries of oligomeric peptide-like molecules1,2,3,4. However, there are no clear-cut rules to guide the design of libraries rich in cell permeable compounds. Furthermore, currently available empirical methods to assess permeability may not accurately reflect true permeability and/or are capable of only modest throughput5,6,7,8,9,10,11,12. We describe here an assay for assessing the relative cell permeability of synthetic molecules in the context of steroid fusions that is capable of high throughput and can be used in any transfectable cell line.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Appella, D.H. et al. Residue-based control of helix shape in beta-peptide oligomers. Nature 387, 381–384 (1997).
Seebach, D., Schaeffer, L., Brenner, M. & Hoyer, D. Design and synthesis of gamma-dipeptide derivatives with submicromolar affinities for human somatostatin receptors. Angew. Chem. Int. Edn. Engl. 42, 776–778 (2003).
Zuckermann, R.N., Kerr, J.M., Kent, S.B.H. & Moos, W.H. Efficient method for the preparation of peptoids [oligo(N-substituted glycines)] by submonomer solid-phase synthesis. J. Am. Chem. Soc. 114, 10646–10647 (1992).
Phillips, S.T., Rezac, M., Abel, U., Kossenjans, M. & Bartlett, P.A. “@-Tides”: The 1,2-Dihydro-3(6H)-pyridinone Unit as a -beta Strand Mimic. J. Am. Chem. Soc. 124, 58–66 (2002).
Artursson, P. & Karlsson, J. Correlation between oral drug absorption in humans and apparent drug permeability coefficients in humans and apparent drug permeability coefficients in human intestinal epithelial (Caco-2) cells. Biochem. Biophys. Res. Commun. 175, 880–885 (1991).
Taylor, E.W., Gibbons, J.A. & Braeckman, R.A. Intestinal absorption screening of mixtures from combinatorial libraries in the Caco-2 model. Pharm. Res. 14, 572–577 (1997).
Goodwin, J.T., Conradi, R.A., Ho, N.F.H. & Burton, P.S. Physicochemical determinants of passive membrane permeability: Role of solute hydrogen-bonding potential and volume. J. Med. Chem. 44, 3721–3729 (2001).
Wang, Z., Hop, C.E.C.A., Leung, K.H. & Pang, J. Determination of in vitro permeability of drug candidates through a Caco-2 cell monolayer by liquid chromatography/tandem mass spectroscopy. J. Mass Spectrom. 35, 71–76 (2000).
Fung, E.N. et al. Higher-throughput screening for Caco-2 permeability utilizing a multiple sprayer liquid chromatography/tandem mass spectrometry system. Rapid Commun. Mass Spectrom. 17, 2147–2152 (2003).
Lundberg, M. & Johansson, M. Is VP22 nuclear homing an artifact? Nat. Biotechnol. 19, 713 (2001).
Richard, J.P. et al. Cell-penetrating peptides: A reevaluation of the mechanism of cellular uptake. J. Biol. Chem. 278, 585–590 (2003).
Belitsky, J.M., Leslie, S.J., Arora, P.S., Beerman, T.A. & Dervan, P.B. Cellular uptake of N-methylpyrrole/N-methylimidazole polyamide-dye conjugates. Bioorg. Med. Chem. 10, 3313–3318 (2002).
Bertorelli, G., Bocchino, V. & Olivieri, D. Heat shock protein interactions with the glucocorticoid receptor. Pulm. Pharmacol. Ther. 11, 7–12 (1998).
Pratt, W.B. & Toft, D.O. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr. Rev. 18, 306–360 (1997).
Brocard, J., Feil, R., Chambon, P. & Metzger, D. A chimeric Cre recombinase inducible by synthetic, but not by natural ligands of the glucocorticoid receptor. Nucleic Acids Res. 26, 4086–4090 (1998).
Braselmann, S., Graninger, P. & Busslinger, M. A selective transcriptional induction system for mammalian cells based on Gal4-estrogen receptor fusion proteins. Proc. Natl. Acad. Sci. USA 90, 1657–1661 (1993).
Wang, Y., O'Malley, B.W., Jr., Tsai, S.Y. & O'Malley, B.W. A regulatory system for use in gene transfer. Proc. Natl. Acad. Sci. USA 91, 8180–8184 (1994).
Rousseau, G.G. & Kirchhoff, J. 17β-Carboxamide steroids are a new class of glucocorticoid antagonists. Nature 279, 158–160 (1979).
Manz, B., Grill, H-J. & Pollow, K. Steroid side-chain modification and receptor affinity: Binding of synthetic derivatives of corticoids to human spleen tumor and rat liver glucocorticoid receptors. J. Steroid Biochem. 17, 335–342 (1982).
Licitra, E.J. & Liu, J.O. A three-hybrid system for detecting small ligand-protein receptor interactions. Proc. Natl. Acad. Sci. USA 93, 12817–12821 (1996).
Alluri, P.G., Reddy, M.M., Bachhawat-Sikder, K., Olivos, H.J. & Kodadek, T. Isolation of protein ligands from large peptoid libraries. J. Am. Chem. Soc. 125, 13995–14004 (2003).
Zuckermann, R.N. et al. Discovery of nanomolar ligands for 7-transmembrane G-protein-coupled receptors from a diverse N-(substituted)glycine peptoid library. J. Med. Chem. 37, 2678–2685 (1994).
Kodadek, T. Development of protein-detecting microarrays and related devices. Trends Biochem. Sci. 27, 295–300 (2002).
Ding, S. et al. Synthetic small molecules that control stem cell fate. Proc. Natl. Acad. Sci. USA 100, 7632–7637 (2003).
Mayer, T.U. et al. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science 286, 971–974 (1999).
Chen, S., Zhang, Q., Wu, X., Schultz, P.G. & Ding, S. Dedifferentiation of lineage-committed cells by a small molecule. J. Am. Chem. Soc. 126, 410–411 (2004).
Peterson, R.T., Link, B.A., Dowling, J.E. & Schreiber, S.L. Small molecule developmental screens reveal the logic and timing of vertebrate development. Proc. Natl. Acad. Sci. USA 97, 12965–12969 (2000).
Burton, P.S., Conradi, R.A., Ho, N.F.H., Hilgers, A.R. & Borchard, R.T. How structural features influence the biomembrane permeability of peptides. J. Pharm. Sci. 85, 1336–1340 (1996).
Goodwin, J.T., Mao, B., Vidmar, T.J., Conradi, R.A. & Burton, P.S. Strategies toward predicting peptide cellular permeability from computed molecular descriptors. J. Pept. Res. 53, 355–369 (1999).
Olivos, H.J., Alluri, P.G., Reddy, M.M., Salony, D. & Kodadek, T. Microwave-assisted solid-phase synthesis of peptoids. Org. Lett. 4, 4057–4059 (2002).
Acknowledgements
We thank John Shelton of the Department of Internal Medicine and Kausar Nadim of the Center for Biomedical Inventions in the UT-Southwestern Medical Center for help with the fluorescence imaging. This work was supported by grants from the National Institutes of Health (P01-DK58398) and the Welch Foundation (I-1299).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Molecular structures of dexamethasone and variousderivatives used in this study. (PDF 700 kb)
Supplementary Fig. 2
Validation of the high throughput assay. (PDF 372 kb)
Supplementary Fig. 3
Evaluation of the cell permeability of the peptoids byfluorescence imaging. (PDF 608 kb)
Supplementary Table 1
Summary of the cell permeability of tetrameric peptoids with different monomer compositions in the high throughput assay. (PDF 54 kb)
Supplementary Table 2
Summary of the parameters from cell permeability and in vitro competition assay for OxDex-tetrameric peptoids carrying different net charges at neutral pH. (PDF 49 kb)
Rights and permissions
About this article
Cite this article
Yu, P., Liu, B. & Kodadek, T. A high-throughput assay for assessing the cell permeability of combinatorial libraries. Nat Biotechnol 23, 746–751 (2005). https://doi.org/10.1038/nbt1099
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt1099
This article is cited by
-
Fluorescent artificial receptor-based membrane assay (FARMA) for spatiotemporally resolved monitoring of biomembrane permeability
Communications Biology (2020)
-
GFP-complementation assay to detect functional CPP and protein delivery into living cells
Scientific Reports (2015)
-
Potent and selective photo-inactivation of proteins with peptoid-ruthenium conjugates
Nature Chemical Biology (2010)
-
A convenient, high-throughput assay for measuring the relative cell permeability of synthetic compounds
Nature Protocols (2007)