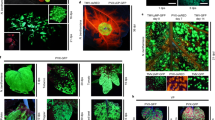
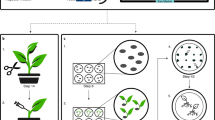
Abstract
Plant biotechnology relies on two approaches for delivery and expression of heterologous genes in plants: stable genetic transformation and transient expression using viral vectors. Although much faster, the transient route is limited by low infectivity of viral vectors carrying average-sized or large genes. We have developed constructs for the efficient delivery of RNA viral vectors as DNA precursors and show here that Agrobacterium–mediated delivery of these constructs results in gene amplification in all mature leaves of a plant simultaneously (systemic transfection). This process, called 'magnifection', can be performed on a large scale and with different plant species. This technology combines advantages of three biological systems (the transfection efficiency of A. tumefaciens, the high expression yield obtained with viral vectors, and the post-translational capabilities of a plant), does not require genetic modification of plants and is faster than other existing methods.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Porta, C. & Lomonossoff, G.P. Viruses as vectors for the expression of foreign sequences in plants. Biotechnol. Genet. Eng. Rev. 19, 245–291 (2002).
Pogue, G.P., Lindbo, J.A., Garger, S.J. & Fitzmaurice, W.P. Making an ally from an enemy: plant virology and the new agriculture. Annu. Rev. Phytopathol. 40, 45–74 (2002).
Gleba, Y., Marillonnet, S. & Klimyuk, V. Engineering viral expression vectors for plants: the 'full virus' and the 'deconstructed virus' strategies. Curr. Opin. Plant Biol. 7, 182–188 (2004).
Mallory, A.C. et al. The amplicon-plus system for high-level expression of transgenes in plants. Nat. Biotechnol. 20, 622–625 (2002).
Marillonnet, S. et al. In planta engineering of viral RNA replicons: efficient assembly by recombination of DNA modules delivered by Agrobacterium . Proc. Natl. Acad. Sci. USA 101, 6852–6857 (2004).
Reed, R. & Hurt, E. A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell 108, 523–531 (2002).
Hebsgaard, S.M. et al. Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res. 24, 3439–3452 (1996).
Voinnet, O., Rivas, S., Mestre, P. & Baulcombe, D. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33, 949–956 (2003).
Kapila, J., De Rycke, R., Van Montagu, M. & Angenon, G. An Agrobacterium-mediated transient gene expression system for intact leaves. Plant Sci. 122, 101–108 (1997).
Turpen, T.H., Turpen, A.M., Weinzettl, N., Kumagai, M.H. & Dawson, W.O. Transfection of whole plants from wounds inoculated with Agrobacterium tumefaciens containing cDNA of tobacco mosaic virus. J. Virol. Methods 42, 227–239 (1993).
Johansen, I.E. Intron insertion facilitates amplification of cloned virus cDNA in Escherichia coli while biological activity is reestablished after transcription in vivo . Proc. Natl. Acad. Sci. USA 93, 12400–12405 (1996).
Yang, S.J. et al. Construction of full-length cDNA clones of lettuce mosaic virus (LMV) and the effects of intron-insertion on their viability in Escherichia coli and on their infectivity to plants. Arch. Virol. 143, 2443–2451 (1998).
Lopez-Moya, J.J. & Garcia, J.A. Construction of a stable and highly infectious intron-containing cDNA clone of plum pox potyvirus and its use to infect plants by particle bombardment. Virus Res. 68, 99–107 (2000).
Racaniello, V.R. & Baltimore, D. Cloned poliovirus complementary DNA is infectious in mammalian cells. Science 214, 916–919 (1981).
Almazan, F. et al. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 97, 5516–5521 (2000).
Lai, M.M. The making of infectious viral RNA: no size limit in sight. Proc. Natl. Acad. Sci. USA 97, 5025–5027 (2000).
Gleba, Y., Klimyu, V. & Marillonnet, S. Magnifection—a new platform for expressing recombinant vaccines in plants. Vaccine 23, 2047–2048 (2005).
Collens, J.I., Lee, D.R., Seeman, A.M. & Curtis, W.R. Development of auxotrophic Agrobacterium tumefaciens for gene transfer in plant tissue culture. Biotechnol. Prog. 20, 890–896 (2004).
Citovsky, V., Zupan, J., Warnick, D. & Zambryski, P. Nuclear localization of Agrobacterium VirE2 protein in plant cells. Science 256, 1802–1805 (1992).
Regensburg-Tuink, A.J. & Hooykaas, P.J. Transgenic N. glauca plants expressing bacterial virulence gene virF are converted into hosts for nopaline strains of A. tumefaciens . Nature 363, 69–71 (1993).
Lee, L.Y., Gelvin, S.B. & Kado, C.I. pSa causes oncogenic suppression of Agrobacterium by inhibiting VirE2 protein export. J. Bacteriol. 181, 186–196 (1999).
Chiu, W. et al. Engineered GFP as a vital reporter in plants. Curr. Biol. 6, 325–330 (1996).
Gallagher, S.R. GUS protocols (ed. Gallagher, S.R.) (Academic Press, Inc., San Diego, 1992).
Acknowledgements
We thank Robert Erwin and Yuri Dorokhov for useful discussions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors are employees of a private company.
Supplementary information
Supplementary Fig. 1
Comparison of the patterns of GFP expression obtained with viral and non-viral vectors. (PDF 39 kb)
Supplementary Fig. 2
Computer prediction of coding and non-coding sequence features in original and synthetic viral vectors. (PDF 515 kb)
Supplementary Table 1
Quantification of the efficiency of initiation of viral replication of the original and modified GFP-expressing viral vectors. (PDF 55 kb)
Rights and permissions
About this article
Cite this article
Marillonnet, S., Thoeringer, C., Kandzia, R. et al. Systemic Agrobacterium tumefaciens–mediated transfection of viral replicons for efficient transient expression in plants. Nat Biotechnol 23, 718–723 (2005). https://doi.org/10.1038/nbt1094
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt1094
This article is cited by
-
Construction of viral-based expression vectors for high-level production of human interferon alpha 2b in plants
Applied Microbiology and Biotechnology (2024)
-
Molecular farming of antimicrobial peptides
Nature Reviews Bioengineering (2023)
-
The emerging role of nanotechnology in plant genetic engineering
Nature Reviews Bioengineering (2023)
-
Viral-based expression cassettes ensure high level production of recombinant green fluorescent protein (GFP) in sweet basil (Ocimum basilicum) plants
Plant Cell, Tissue and Organ Culture (PCTOC) (2023)
-
Tobacco as green bioreactor for therapeutic protein production: latest breakthroughs and optimization strategies
Plant Growth Regulation (2023)