Abstract
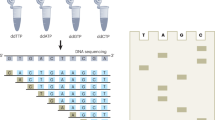
The determination of the complete nucleotide sequence of large DNA molecules is a tedious and time-consuming process. The repetitive nature of the task makes it an ideal subject for automation. We report here a system designed and constructed to perform the chain termination reactions of the Sanger sequencing procedure automatically. It is capable of carrying out these reactions for from five to twelve DNA templates at one time. The system takes 12 minutes to dispense 12 templates, add the reagents and mix them; the complete cycle time, including incubations, is 45 minutes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Maxam, A.M. and Gilbert, W. 1977. A new method for sequencing DNA. Proc. Natl. Acad. Sci. 74: 560–56.
Maxam, A.M. and Gilbert, W. 1980. Sequencing end-labelled DNA with base-specific chemical cleavage, p. 499–560. In: Methods in Enzymology, Vol. 65, Grossman, L. and Moldave, K. (eds.) Academic Press, New York.
Sanger, F., Niklen, S. and Coulson, A.R. 1977. DNA sequencing with chain terminating inhibitors. Proc. Natl. Acad. Sci. 74: 5463–5467.
Gronenborn, B. and Messing, J. 1978. Methylation of single-stranded DNA in vitro introduces new restriction endonuclease cleavage sites. Nature 272: 375–37.
Messing, J. and Vieira, J. 1982. A new pair of M13 vectors for selecting either DNA strand of double-digested restriction fragments. Gene 19: 269–276.
Sanger, F., Coulson, A.R., Hong, G.F., Hill, D.F. and Peterson, G.B. 1982. Nucleotide sequence of bacteriophage DNA. J. Mol. Biol. 162: 729–773.
Baer, R., Bankier, A.T., Biggin, M.D., Deringer, P.L., Farrel, P.J., Gibson, T.J., Hatful, G., Hudson, G.S., Satchwell, S.C., Seguin, C., Tuffnel, P.S. and Barrell, B.G. 1984. DNA sequence and the expression of the B95-8 Epstein-Barr virus genome. Nature 310: 207–211.
Waring, R.B., Davies, R.W., Lee, S., Grisi, E., McPhail Berks, M. and Scazzochio, C. 1982. Mosaic organisation of the apocytochrome b gene of Aspergillus nidulans as revealed by DNA sequencing. Cell 27: 4–11.
Wada, A., Yamamoto, M. and Soeda, E. 1983. Automatic DNA sequencer: Computer-programmed microchemical manipulator for the Maxam-Gilbert sequencing method. Rev. Sci. Instru. 54: 1569–1572.
Deninger, P.L. 1983. Random subcloning of sonicated DNA: application to shotgun DNA sequence analysis. Analyt. Biochem. 129: 216–223.
Staden, R. 1980. A new computer method for the storage and manipulation of DNA gel reading data. Nucleic Acids. Res. 8: 3673–3694.
Staden, R. and McLachlan, A.D. 1982. Codon preference and its use in identifying protein coding regions in long DNA sequences. Nucleic Acids. Res. 10: 141–156.
Martin, W.J. U.K. Patent application 8500294 of 7th January 1985.
Barnes, W.M., Bevan, M. and Son, P.H. 1980. Kilo-sequencing: creation of an ordered nest of asymmetric deletions across a larger target sequence carried on phage M13, p. 98–122. In: Methods in Enzymology, Vol. 10. Grossman, L. and Moldave, K. (eds.), Academic Press, New York.
Biggin, M.D., Gibson, T.J. and Hong, G.F. 1980. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc. Natl. Acad. Sci. 80: 3963–3965.
Charpak, G., Melchart, G., Petersen, G. and Sauli, F. 1981. The Multistep chamber as a high accuracy localization device for B-chromatography and slow-neutron imaging. IEEE Trans. Nuc. Sc. 28: 849–851.
Scott, B.J., Bateman, J.E. and Bradwell, A.R. 1982. The detection of tritium labelled ligands and their carrier proteins using a multiwire proportional counter. Anal. Biochem. 123: 1–10.
Beck, S. and Pohl, F.M. 1984. DNA sequencing with direct blotting electrophoresis. EMBO. J. 3: 2905–2909.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Martin, W., Warmington, J., Galinski, B. et al. Automation of DNA Sequencing: A System to Perform the Sanger Dideoxysequencing Reactions. Nat Biotechnol 3, 911–915 (1985). https://doi.org/10.1038/nbt1085-911
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nbt1085-911
This article is cited by
-
Optimization of Parameters in a DNA Sequenator Using Fluorescence Detection
Nature Biotechnology (1988)
-
Automation of DNA Sequencing Reactions and Related Techniques: A Workstation for Micromanipulation of Liquids
Bio/Technology (1988)
-
Automating the Purification and Isolation of Synthetic DNA
Nature Biotechnology (1987)
-
Automated DNA Sequencing: Progress and Prospects
Nature Biotechnology (1986)