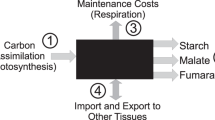
Abstract
The study of the metabolite complement of biological samples, known as metabolomics, is creating large amounts of data, and support for handling these data sets is required to facilitate meaningful analyses that will answer biological questions. We present a data model for plant metabolomics known as ArMet (architecture for metabolomics). It encompasses the entire experimental time line from experiment definition and description of biological source material, through sample growth and preparation to the results of chemical analysis. Such formal data descriptions, which specify the full experimental context, enable principled comparison of data sets, allow proper interpretation of experimental results, permit the repetition of experiments and provide a basis for the design of systems for data storage and transmission. The current design and example implementations are freely available (http://www.armet.org/). We seek to advance discussion and community adoption of a standard for metabolomics, which would promote principled collection, storage and transmission of experiment data.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Bob Crimi

Bob Crimi

Bob Crimi
Similar content being viewed by others
References
Quackenbush, J. Data standards for 'omic' science. Nat. Biotechnol. 22, 613–614 (2004).
Brazma, A. et al. Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nat. Genet. 29, 365–371 (2001).
Spellman, P.T. et al. Design and implementation of microarray gene expression markup language (MAGE-ML). Genome Biol. 3, research0046.0041–0046.0049 (2002).
Brazma, A. et al. ArrayExpress—a public repository for microarray gene expression data at the EBI. Nucleic Acids Res. 31, 68–71 (2003).
Killion, P.J., Sherlock, G. & Iyer, V.R. The Longhorn Array Database (LAD): An open-source, MIAME compliant implementation of the Stanford Microarray database (SMD). BMC Bioinformatics 4, 32 (2003). http://biomedcentral.com/1471-2105/4
Editorial. Microarray standards at last. Nature 419, 323 (2002).
Glueck, S.B. & Dzau, V.J. Our new requirement for MIAME standards. Physiol. Genomics 13, 1–2 (2003).
Oliver, S. On the MIAME standards and central repositories, of microarray. Comp. Funct. Genomics 4, 1 (2003).
Taylor, C.F. et al. A systematic approach to modeling, capturing, and disseminating proteomics experimental data. Nat. Biotechnol. 21, 247–254 (2003).
Booch, G., Rumbaugh, J. & Jacobson, I. The Unified Modeling Language User Guide (Addison-Wesley, Reading, MA, 1999).
Orchard, S., Hermjakob, H. & Apweiler, R. The proteomics standards initiative. Proteomics 3, 1374–1376 (2003).
Orchard, S. et al. Common interchange of standards for proteomics data: public availability of tools and schema. Proteomics 4, 490–491 (2004).
Fiehn, O. Combining genomics, metabolome analysis, and biochemical modelling to understand metabolic networks. Comp. Funct. Genomics 2, 155–168 (2001).
Fiehn, O. Metabolomics—the link between genotypes and phenotypes. Plant Mol. Biol. 48, 155–171 (2002).
Fiehn, O. & Weckwerth, W. Deciphering metabolic networks. Eur. J. Biochem. 270, 579–588 (2003).
Goodacre, R., Vaidyanathan, S., Dunn, W.B., Harrigan, G.G. & Kell, D.B. Metabolomics by numbers:acquiring and understanding global metabolite data. Trends Biotechnol. 22, 245–252 (2004).
Harrigan, G.G. & Goodacre, R. (eds.) Metabolic Profiling: Its Role in Biomarker Discovery and Gene Function Analysis (Kluwer Academic Publishers, Boston, 2003).
Mendes, P. Emerging bioinformatics for the metabolome. Brief. Bioinform. 3, 134–145 (2002).
Oliver, S.G., Winson, M.K., Kell, D.B. & Baganz, F. Systematic functional analysis of the yeast genome. Trends Biotechnol. 16, 373–378 (1998).
Raamsdonk, L.M. et al. A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations. Nat. Biotechnol. 19, 45–50 (2001).
Roessner, U. et al. Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13, 11–29 (2001).
Roessner, U., Wagner, C., Kopka, J., Trethewey, R.N. & Willmitzer, L. Simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J. 23, 131–142 (2000).
Sumner, L.W., Mendes, P. & Dixon, R.A. Plant metabolomics: large-scale phytochemistry in the functional genomics era. Phytochemistry 62, 817–836 (2003).
Weckwerth, W. Metabolomics in systems biology. Annu. Rev. Plant Biol. 54, 669–689 (2003).
Hall, R. et al. Plant metabolomics: the missing link in functional genomics strategies (Meeting report). Plant Cell 14, 1437–1440 (2002).
van der Greef, J., van der Heiiden, R. & Verheij, E.R. The role of mass spectrometry in system biology: data processing and identification strategies in metabolomics. in Advances in Mass Spectrometry, vol. 16. (eds. Ashcroft, A.E., Brenton, G. & Monaghan, J.J.) 145–165 (Elsevier, Amsterdam, 2004).
Kanehisa, M. & Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30 (2000).
Krieger, C.J. et al. MetaCyc: a multiorganism database of metabolic pathways and enzymes. Nucleic Acids Res. 32, D438–D442 (2004).
Mueller, L.A., Zhang, P. & Rhee, S.Y. AraCyc: A biochemical pathway database for Arabidopsis. Plant Physiol. 132, 453–460 (2003).
Harris, M.A. et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 32 Special Issue, D258–D261 (2004).
Nicholson, J.K., Lindon, J.C. & Homes, E. 'Metabonomics': understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 29, 1181–1189 (1999).
Bino, R.J. et al. Potential of metabolomics as a functional genomics tool. Trends Plant Sci. 9, 418–425 (2004).
Stein, S.E. & Scott, D.R. Optimization and testing of mass spectral library search algorithms for compound identification. J. Am. Soc. Mass Spectrom. 5, 859–866 (1994).
McLafferty, F.W., Zhang, M-Y., Stauffer, D.B. & Loh, S.Y. Comparison of algorithms and databases for matching unknown mass spectra. J. Am. Soc. Mass Spectrom. 9, 92–95 (1998).
Xirasagar, S. et al. CEBS object model for systems biology data, SysBio-OM. Bioinformatics 20, 2004–2015 (2004).
Lampen, P. et al. An extension to the JCAMP-DX standard file format, JCAMP-DX V.5.01 (IUPAC Recommendations 1999). Pure Appl. Chem. 71, 1549–1556 (1999).
Griffiths, P.R. & de Haseth, J.A. Fourier Transform Infrared Spectrometry, vol. 83 (John Wiley & Sons, Chichester, England, 1986).
Allen, J.K. et al. High-throughput classification of yeast mutants for functional genomics using metabolic footprinting. Nat. Biotechnol. 21, 692–696 (2003).
Smedsgaard, J. Terverticillate penicillia studied by direct electrospray mass spectrometric profiling of crude extracts: II. Database and identification. Biochemical Systematics and Ecology 25, 65–71 (1997).
Smedsgaard, J. & Frisvad, J.C. Using direct electrospray mass spectrometry in taxonomy and secondary metabolite profiling of crude fungal extracts. J Microbiol. Methods 25, 5–17 (1996).
Acknowledgements
The authors gratefully acknowledge the United Kingdom Food Standards Agency (under the G02006 project), the United Kingdom Biotechnology and Biological Sciences Research Council (particularly under the HiMet project) and the United Kingdom Engineering and Physical Sciences Research Council for support of their work in metabolomics. We would also like to thank personnel at the United Kingdom Institute of Grassland and Environmental Research and delegates at the 1st, 2nd and 3rd International Plant Metabolomics Conferences for many useful discussions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Jenkins, H., Hardy, N., Beckmann, M. et al. A proposed framework for the description of plant metabolomics experiments and their results. Nat Biotechnol 22, 1601–1606 (2004). https://doi.org/10.1038/nbt1041
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt1041
This article is cited by
-
Biological big-data sources, problems of storage, computational issues, and applications: a comprehensive review
Knowledge and Information Systems (2024)
-
Impact of low-intensity pulsed ultrasound on transcription and metabolite compositions in proliferation and functionalization of human adipose-derived mesenchymal stromal cells
Scientific Reports (2020)
-
Towards a standardized bioinformatics infrastructure for N- and O-glycomics
Nature Communications (2019)
-
Compliance with minimum information guidelines in public metabolomics repositories
Scientific Data (2017)
-
Introduction to metabolomics and its applications in ophthalmology
Eye (2016)