Abstract
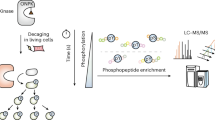
To study the global dynamics of phosphotyrosine-based signaling events in early growth factor stimulation, we developed a mass spectrometric method that converts temporal changes to differences in peptide isotopic abundance. The proteomes of three cell populations were metabolically encoded with different stable isotopic forms of arginine. Each population was stimulated by epidermal growth factor for a different length of time, and tyrosine-phosphorylated proteins and closely associated binders were affinity purified. Arginine-containing peptides occurred in three forms, which were quantified; we then combined two experiments to generate five-point dynamic profiles. We identified 81 signaling proteins, including virtually all known epidermal growth factor receptor substrates, 31 novel effectors and the time course of their activation upon epidermal growth factor stimulation. Global activation profiles provide an informative perspective on cell signaling and will be crucial to modeling signaling networks in a systems biology approach.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Pawson, T. & Nash, P. Assembly of cell regulatory systems through protein interaction domains. Science 300, 445–452 (2003).
Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 103, 211–225 (2000).
Blume-Jensen, P. & Hunter, T. Oncogenic kinase signalling. Nature 411, 355–365 (2001).
Ficarro, S.B. et al. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol. 20, 301–305 (2002).
Salomon, A.R. et al. Profiling of tyrosine phosphorylation pathways in human cells using mass spectrometry. Proc. Natl. Acad. Sci. USA 100, 443–448 (2003).
Gygi, S.P. et al. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 17, 994–999 (1999).
Lill, J. Proteomic tools for quantitation by mass spectrometry. Mass Spectrom. Rev. 22, 182–194 (2003).
Aebersold, R. & Mann, M. Mass spectrometry-based proteomics. Nature 422, 198–207 (2003).
Ranish, J.A. et al. The study of macromolecular complexes by quantitative proteomics. Nat. Genet. 33, 349–355 (2003).
Blagoev, B. et al. A proteomics strategy to elucidate functional protein-protein interactions applied to EGF signaling. Nat. Biotechnol. 21, 315–318 (2003).
Ong, S.E. et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 (2002).
Ong, S.E., Kratchmarova, I. & Mann, M. Properties of 13C-substituted arginine in stable isotope labeling by amino acids in cell culture (SILAC). J. Proteome Res. 2, 173–181 (2003).
Pandey, A. et al. Identification of a novel immunoreceptor tyrosine-based activation motif-containing molecule, STAM2, by mass spectrometry and its involvement in growth factor and cytokine receptor signaling pathways. J. Biol. Chem. 275, 38633–38639 (2000).
Pandey, A. et al. Analysis of receptor signaling pathways by mass spectrometry: identification of vav-2 as a substrate of the epidermal and platelet- derived growth factor receptors. Proc. Natl. Acad. Sci. USA 97, 179–184 (2000).
Pandey, A. et al. Cloning of a novel phosphotyrosine binding domain containing molecule, Odin, involved in signaling by receptor tyrosine kinases. Oncogene 21, 8029–8036 (2002).
Waterman, H. & Yarden, Y. Molecular mechanisms underlying endocytosis and sorting of ErbB receptor tyrosine kinases. FEBS Lett. 490, 142–152 (2001).
Dikic, I. & Giordano, S. Negative receptor signalling. Curr. Opin. Cell Biol. 15, 128–135 (2003).
Bache, K.G., Raiborg, C., Mehlum, A. & Stenmark, H. STAM and Hrs are subunits of a multivalent ubiquitin-binding complex on early endosomes. J. Biol. Chem. 278, 12513–12521 (2003).
Dong, C., Waters, S.B., Holt, K.H. & Pessin, J.E. SOS phosphorylation and disassociation of the Grb2-SOS complex by the ERK and JNK signaling pathways. J. Biol. Chem. 271, 6328–6332 (1996).
De Corte, V. et al. Gelsolin-induced epithelial cell invasion is dependent on Ras-Rac signaling. EMBO J. 21, 6781–6790 (2002).
Riggins, R.B., Quilliam, L.A. & Bouton, A.H. Synergistic promotion of c-Src activation and cell migration by Cas and AND-34/BCAR3. J. Biol. Chem. 278, 28264–28273 (2003).
Brinkman, A., van der Flier, S., Kok, E.M. & Dorssers, L.C. BCAR1 a human homologue of the adapter protein p130Cas, and antiestrogen resistance in breast cancer cells. J. Natl. Cancer Inst. 92, 112–120 (2000).
van Agthoven, T. et al. Identification of BCAR3 by a random search for genes involved in antiestrogen resistance of human breast cancer cells. EMBO J. 17, 2799–2808 (1998).
Ostareck-Lederer, A. et al. c-Src-mediated phosphorylation of hnRNP K drives translational activation of specifically silenced mRNAs. Mol. Cell. Biol. 22, 4535–4543 (2002).
Bach, I. The LIM domain: regulation by association. Mech. Dev. 91, 5–17 (2000).
Wu, R. et al. Specificity of LIM domain interactions with receptor tyrosine kinases. J. Biol. Chem. 271, 15934–15941 (1996).
Seykora, J.T., Mei, L., Dotto, G.P. & Stein, P.L. 'Srcasm: a novel Src activating and signaling molecule. J. Biol. Chem. 277, 2812–2822 (2002).
Lohi, O., Poussu, A., Mao, Y., Quiocho, F. & Lehto, V.P. VHS domain–a longshoreman of vesicle lines. FEBS Lett. 513, 19–23 (2002).
Trompouki, E. et al. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature 424, 793–796 (2003).
Kovalenko, A. et al. The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature 424, 801–805 (2003).
Brummelkamp, T.R., Nijman, S.M., Dirac, A.M. & Bernards, R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature 424, 797–801 (2003).
Wiley, H.S., Shvartsman, S.Y. & Lauffenburger, D.A. Computational modeling of the EGF-receptor system: a paradigm for systems biology. Trends Cell Biol. 13, 43–50 (2003).
Shevchenko, A., Wilm, M., Vorm, O. & Mann, M. Mass spectrometric sequencing of proteins from silver stained polyacrylamide gels. Anal. Chem. 68, 850–858 (1996).
Rappsilber, J., Ishihama, Y. & Mann, M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 (2003).
Acknowledgements
We thank other members of our laboratory for help and fruitful discussions. Peter Mortensen is acknowledged for help with the bioinformatic analysis and Carmen de Hoog and Leonard Foster for critical reading of the manuscript. Work in the Center for Experimental BioInformatics (CEBI) is supported by a generous grant by the Danish National Research foundation and by the 6th Framework Program of the European Union (Interaction Proteome).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Variability of kinetic data. (PDF 44 kb)
Supplementary Table 1
Proteins activated upon EGF stimulation. (PDF 111 kb)
Rights and permissions
About this article
Cite this article
Blagoev, B., Ong, SE., Kratchmarova, I. et al. Temporal analysis of phosphotyrosine-dependent signaling networks by quantitative proteomics. Nat Biotechnol 22, 1139–1145 (2004). https://doi.org/10.1038/nbt1005
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt1005
This article is cited by
-
Genome-wide analysis of DNA methylation and risk of cardiovascular disease in a Chinese population
BMC Cardiovascular Disorders (2021)
-
ALKBH3 partner ASCC3 mediates P-body formation and selective clearance of MMS-induced 1-methyladenosine and 3-methylcytosine from mRNA
Journal of Translational Medicine (2021)
-
A translational program that suppresses metabolism to shield the genome
Nature Communications (2020)
-
Coiled-coil domain containing 50-V2 protein positively regulates neurite outgrowth
Scientific Reports (2020)
-
Global quantitative analysis of the human brain proteome and phosphoproteome in Alzheimer’s disease
Scientific Data (2020)