Abstract
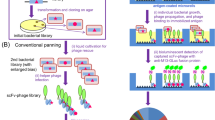
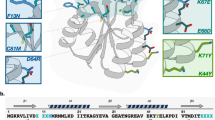
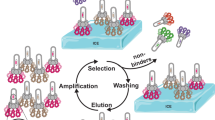
We describe a method for selecting aggregation-resistant proteins by heat denaturation. This is illustrated with antibody heavy chain variable domains (dAbs), which are prone to aggregate1,2. The dAbs were displayed multivalently at the infective tip of filamentous bacteriophage, and heated transiently to induce unfolding and to promote aggregation of the dAbs. After cooling, the dAbs were selected for binding to protein A (a ligand common to these folded dAbs). Phage displaying dAbs that unfold reversibly were thereby enriched with respect to those that do not. From a repertoire of phage dAbs, six dAbs were characterized after selection; they all resisted aggregation, and were soluble, well expressed in bacteria and could be purified in good yields. The method should be useful for making aggregation-resistant proteins and for helping to identify features that promote or prevent protein aggregation, including those responsible for misfolding diseases3,4.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Change history
15 August 2004
appended corrigendum pdf to AOP PDF; corrected online date will appear in print
Notes
*Note: In the PDF of the version of this article originally published online, approximately 600 words of text was omitted, including the end of paragraph three through the first half of paragraph eight. This mistake has been corrected for the print version of this article.
References
Ward, E.S., Güssow, D., Griffiths, A.D., Jones, P.T. & Winter, G. Binding activities of a repertoire of single immunoglobulin variable domains secreted from Escherichia coli. Nature 341, 544–546 (1989).
Ewert, S., Cambillau, C., Conrath, K. & Plückthun, A. Biophysical properties of camelid VHH domains compared to those of human VH3 domains. Biochemistry 41, 3628–3636 (2002).
Dobson, C.M. Protein misfolding, evolution and disease. Trends Biochem. Sci. 24, 329–332 (1999).
Rochet, J.C. & Lansbury P.T., Jr. Amyloid fibrillogenesis: themes and variations. Curr. Opin. Struct. Biol. 10, 60–68 (2000).
Dumoulin, M. et al. Single-domain antibody fragments with high conformational stability. Protein Sci. 11, 500–515 (2002).
Perez, J.M. et al. Thermal unfolding of a llama antibody fragment: a two-state reversible process. Biochemistry 40, 74–83 (2001).
van der Linden, R.H. et al. Comparison of physical chemical properties of llama VHH antibody fragments and mouse monoclonal antibodies. Biochim. Biophys. Acta 1431, 37–46 (1999).
Jespers, L., Schon, O., James, L.C., Veprintsev, D. & Winter, G. Crystal structure of HEL4, a soluble, refoldable human VH single domain with a germ-line scaffold. J. Mol. Biol. 337, 893–903 (2004).
McCafferty, J., Griffiths, A.D., Winter, G. & Chiswell, D.J. Phage antibodies: filamentous phage displaying antibody variable domains. Nature 348, 552–554 (1990).
Holliger, P., Riechmann, L. & Williams, R.L. Crystal structure of the two N-terminal domains of g3p from filamentous phage fd at 1.9 Å: evidence for conformational lability. J. Mol. Biol. 288, 649–657 (1999).
Wilkinson, D.L. & Harrison, R.G. Predicting the solubility of recombinant proteins in Escherichia coli. Biotechnology (NY) 9, 443–448 (1991).
Chiti, F. et al. Studies of the aggregation of mutant proteins in vitro provide insights into the genetics of amyloid diseases. Proc. Natl. Acad. Sci. USA 99, 16419–16426 (2002).
Ewert, S., Huber, T., Honegger, A. & Plückthun, A. Biophysical properties of human antibody variable domains. J. Mol. Biol. 325, 531–553 (2003).
Sepulveda, J., Jin, H., Sblattero, D., Bradbury, A. & Burrone, O.R. Binders based on dimerised immunoglobulin VH domains. J. Mol. Biol. 333, 355–365 (2003).
Arbabi Ghahroudi, M., Desmyter, A., Wyns, L., Hamers, R. & Muyldermans, S. Selection and identification of single domain antibody fragments from camel heavy-chain antibodies. FEBS Lett. 414, 521–526 (1997).
Kristensen, P. & Winter, G. Proteolytic selection for protein folding using filamentous bacteriophages. Fold. Des. 3, 321–328 (1998).
Sieber, V., Plückthun, A. & Schmid, F.X. Selecting proteins with improved stability by a phage-based method. Nat. Biotechnol. 16, 955–960 (1998).
Martin, A., Sieber, V. & Schmid, F.X. In vitro selection of highly stabilized protein variants with optimized surface. J. Mol. Biol. 309, 717–726 (2001).
Shusta, E.V., Holler, P.D., Kieke, M.C., Kranz, D.M. & Wittrup, K.D. Directed evolution of a stable scaffold for T-cell receptor engineering. Nat. Biotechnol. 18, 754–759 (2000).
Jung, S., Honegger, A. & Plückthun, A. Selection for improved protein stability by phage display. J. Mol. Biol. 294, 163–180 (1999).
Tomlinson, I.M., Walter, G., Marks, J.D., Llewelyn, M.B. & Winter, G. The repertoire of human germline VH sequences reveals about fifty groups of VH segments with different hypervariable loops. J. Mol. Biol. 227, 776–798 (1992).
Wörn, A. & Plückthun, A. Stability engineering of antibody single-chain Fv fragments. J. Mol. Biol. 305, 989–1010 (2001).
Muyldermans, S., Atarhouch, T., Saldanha, J., Barbosa, J.A. & Hamers, R. Sequence and structure of VH domain from naturally occurring camel heavy chain immunoglobulins lacking light chains. Protein Eng. 7, 1129–1135 (1994).
Desmyter, A. et al. Crystal structure of a camel single-domain VH antibody fragment in complex with lysozyme. Nat. Struct. Biol. 3, 803–811 (1996).
Waldo, G.S. Genetic screens and directed evolution for protein solubility. Curr. Opin. Chem. Biol. 7, 33–38 (2003).
Kabat, E.A. National Institutes of Health (US) & Columbia University. Sequences of proteins of immunological interest, edn. 5 (US Dept. of Health and Human Services Public Health Service, National Institutes of Health, Bethesda, MD, 1991).
Pace, C.N. & Scholtz, J.M. in Protein Structure, A Practical Approach, edn. 2 (ed. Creighton, T.E.) 299–321 (Oxford University Press, New York, 1997).
Myers, J.K., Pace, C.N. & Scholtz, J.M. Denaturant m values and heat capacity changes: relation to changes in accessible surface areas of protein unfolding. Protein Sci. 4, 2138–2148 (1995).
Chothia, C. et al. Structural repertoire of the human VH segments. J. Mol. Biol. 227, 799–817 (1992).
Acknowledgements
We thank John Berriman for expert assistance with the electron microscopy. While working at the Laboratory of Molecular Biology, L.J. and O.S. were funded by Domantis Limited (Cambridge, UK) under a collaborative research program with the Medical Research Council.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
G.W. is a founder, shareholder and director of Domantis.
Rights and permissions
About this article
Cite this article
Jespers, L., Schon, O., Famm, K. et al. Aggregation-resistant domain antibodies selected on phage by heat denaturation. Nat Biotechnol 22, 1161–1165 (2004). https://doi.org/10.1038/nbt1000
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt1000
This article is cited by
-
Non-specificity as the sticky problem in therapeutic antibody development
Nature Reviews Chemistry (2022)
-
Local environment effects on charged mutations for developing aggregation-resistant monoclonal antibodies
Scientific Reports (2020)
-
An in vivo platform to select and evolve aggregation-resistant proteins
Nature Communications (2020)
-
The biodistribution and clearance of AlbudAb, a novel biopharmaceutical medicine platform, assessed via PET imaging in humans
EJNMMI Research (2019)
-
Selection and screening strategies in directed evolution to improve protein stability
Bioresources and Bioprocessing (2019)