Abstract
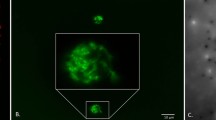
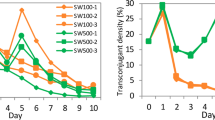
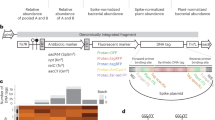
A sensitive, selectable marker system has been developed for fluorescent pseudomonads based on expression of the E. coli lac operon genes encoding β–galactosidase and lactose permease. Examination of over 500 fluorescent Pseudomonas isolates from soil samples revealed the constant absence of the o–nitrophenylgalactoside (ONPG) positive phenotype, and the inability to utilize lactose as a sole carbon source. Based on these observations broad host–range plasmids containing E. coli lacZ and lacY genes were constructed and stably introduced into native P. fluorescens strains to confer the ability to cleave the chromogenic substrate X–Gal and to grow on minimal lactose media. This marker system enabled the detection of Lac+ transformants at a sensitivity of <10 CFU/g soil.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Office of Technology Assessment. 1983. Commercial biotechnology: An international analysis. Washington, D.C.
Gillett, J.W., Levin, S.A., Harwell, M.A., Andow, D.A., Alexander, M. and Stern, A.M. 1984. Potential impacts of environmental release of biotechnology products: Assessment, regulation, and research needs. Ecosystems Research Center, Cornell University, Ithaca, N.Y.
Brill, W.J. 1985. Safety concerns and genetic engineering in agriculture. Science 227:381–384.
Covello, V.T. and Fiksel, J.R. 1985. The suitability and applicability of risk assessment methods for environmental applications of biotechnology. National Science Foundation, Report No. NSF/PRA 8502286, Washington, D.C.
Schroth, M.N., Thomson, S.V., and Moller, W.J. 1979. Streptomycin resistance in Erwinia amylovora. Phytopathology 69:565–568.
Burr, T.J., Schroth, M.N. and Suslow, T. 1978. Increased potato yield by treatment of seedpieces with specific strains of Pseudomonas fluorescens and P. putida. Phytopathology 68:1377–1383.
Kloepper, J.W., Schroth, M.N., and Miller, T.D. 1980. Effects of rhizosphere colonization by plant growth-promoting rhizobacteria on potato plant development and yield. Phytopathology 70:1078–1082.
Suslow, T.V. and Schroth, M.N. 1982. Rhizobacteria of sugar beets: Effects of seed application and root colonization on yield. Phytopathology 72:199–206.
Hemming, B.C. and Drahos, D.J. 1984. β-Galactosidase, a selectable non-antibiotic marker for fluorescent pseudomonads. J. Cellular Biochemistry Supp. 8b:252.
Kalnins, A., Otto, K., Ruther, U., and Muller-Hill, B. 1983. Sequence of the lacZ gene of Escherichia coli. EMBO Journal 2:593–597.
Buchel, D.E., Gronenborn, B., and Muller-Hill, B. 1980. Sequence of the lactose permease gene. Nature 283:541–545.
Zabin, I. and Fowler, A.V. 1978. β-Galactosidase, the lactose permease protein, and thiogalactoside transacetylase, p. 89–121. In: The Operon. J. Miller and W. Resnikoff, (eds.) Cold Spring Harbor Laboratory, New York.
Teather, R.M., Bramhall, J., Riede, I., Wright, J.K., Furst, M., Aichele, G., Wilhelm, R., and Overath, P. 1980. Lactose carrier protein of Escherichia coli. Eur. J. Biochem 108:223–231.
Mieschendahl, M., Buchel, D., Bocklage, H., and Muller-Hill, B. 1981. Mutations in the lacY gene of Escherichia coli define functional organization of lactose permease. Proc. Natl. Acad. Sci. USA 78:7652–7656.
Weller, D.M. 1983. Colonization of wheat roots by a fluorescent pseudomonad suppressive to take-all. Phytopathology 73:1548–1553.
Weller, D.M. and Cook, R.J. 1982. Pseudomonads from take-all conducive and suppressive soils. Phytopathology 72:264.
Weller, D.M. and Cook, R.J. 1983. Suppression of take-all of wheat by seed-treatment with fluorescent pseudomonads. Phytopathology 73:463–469.
Ballard, R.W., Palleroni, N.J., Doudoroff, M., Stanier, R.Y., and Mandel, M. 1970. Taxonomy of the aerobic pseudomonads: Pseudomonas cepacia, P. marginala, P. alliicola and P. caryophylli. J. Gen. Microbiol. 60:199–214.
Palleroni, N.J. and Doudoroff, M. 1972. Some properties and taxonomic subdivisions of the genus Pseudomonas. Ann. Rev. Phytopathology 10:73–100.
Stanier, R.Y., Palleroni, N.J., and Doudoroff, M. 1966. The aerobic pseudomonads: a taxonomic study. J. Gen. Microbiol. 43:159–271.
Okamura, Y., Shoda, M., and Shigezo, U. 1983. Activity and synthesis of β-galactosidase in various lactose utilizing bacteria. Agric. Biol. Chem. 47:1133–1134.
Analytab Products, Ayerst Laboratories, Inc. 1982. Analytical profile index, enterobacteriaceae and other gram negative bacteria. Plain-view, New York.
Hayward, A.C. 1978. Occurrence of glycoside hydrolases in plant pathogenic and related bacteria. J. Appl. Bacteriol. 43:407–412.
Bagdasarian, M., Lurz, R., Ruckert, B., Franklin, F.C.H., Bagdasarian, M.M., Frey, J., and Timmis, K.N. 1981. Specific-purpose plasmid cloning vectors II. Broad host-range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene 16:237–247.
Casadaban, M.J., Chou, J., and Cohen, S.N. 1980. In vitro gene fusions that join an enzymatically active β-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J. Bacteriol. 143:971–980.
Maniatis, T., Fritsch, E., and Sambrook, J. 1982. Molecular Cloning, a Laboratory Manual. Cold Spring Harbor Laboratory, New York.
Figurski, D.H. and Helinski, D.R. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648–1652.
Kawalek, M. and Schaad, N.W. 1985. β-galactosidase as a metabolic marker in Xanthomonas campestris pv. campestris. Phytopathology 75:1326.
Miller, J. 1972. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory, N.Y.
Newman, M.J., Foster, D.L., Wilson, T.H., Kaback, H.R. 1981. Purification and reconstitution of functional lactose carrier from E. coli. J. Biol. Chem. 256:11804–11808.
Renart, J., Reiser, J., and Stark, G.R. 1979. Transfer of proteins from gels to diazobenzyloxymethyl-paper and detection with antisera: A method for studying antibody specificity and antigen structure. Proc. Natl. Acad. Sci. USA. 76:3116–3120.
Gillaspie, A.G., Sasser, M., and Davis, M.J. 1984. Fatty acid profiles of bacteria causing ratoon stunting disease (RSD) of sugarcane and bermudagrass stunting disease (BSD). Phytopathology 74:880.
Drahos, D., Brackin, M.J., and Barry, G. 1985. Bacterial strain identification by comparative analysis of chromosomal DNA restriction patterns. Phytopathology 75:1381.
Krivi, G.G. and Rowold, E. 1984. Monoclonal antibodies to bovine somatotropin: Immunoadsorbent reagents for mammalian somatotropins. Hybridoma 3:151–162.
Cohen, G.N. and Monad, J. 1957. Bacterial permeases. Bacteriol. Rev. 21:169–194.
Marin, A. and Marshall, R.T. 1983. Characterization of glycosidases produced by Pseudomonas fluorescens 26. J. Food Protection 46:676–680.
Gorbach, S.L. 1971. Intestinal microflora. Gastroenterology 60:1110–1131.
Stotzky, G. and Babich, H. 1984. Fate of genetically-engineered microbes in natural environments. Recombinant DNA Technical Bulletin 7:163–188.
Cole, M.A. and Elkan, G.H. 1979. Multiple antibiotic resistance in Rhizobium japonicum. Appl. Environ. Microbiol. 37:867–870.
Kelch, W.J. and Lee, J.S. 1978. Antibiotic resistance patterns of gram-negative bacteria isolated from environmental sources. Appl. Environ. Microbiol. 36:450–456.
Kloepper, J.W., Leong, J., Teintze, M., and Schroth, M.N. 1980. Pseudomonas siderophores: A mechanism explaining disease-suppressive soils. Current Microbiology 4:317–320.
Godwin, D. and Slater, J.H. 1979. The influence of the growth environment on the stability of a drug resistance plasmid in Escherichia coli K-12. J. Gen. Microbiol. 111:201–210.
Kloepper, J.W. and Scroth, M.N. 1981. Relationship of in vitro antibiosis of plant growth-promoting rhizobacteria to plant growth and the displacement of root microflora. Phytopathology 71:1020–1024.
Cook, R.J. and Baker, K.F. 1983. The nature and practice of biological control of plant pathogens. The American Phytopathological Society, St. Paul, Minnesota.
Chopra, I., Shales, S.W., Ward, M.W., and Wallace, L.J. 1981. Reduced expression of Tn10-mediated tetracycline resistance in Escherichia coli containing more than one copy of the transposon. J. Gen. Microbiol. 126:45–54.
Levy, S.B. 1984. Resistance to the tetracyclines, p. 191–240. In: Antimicrobial Drug Resistance. L. E. Bryan (ed.) Academic Press, Inc. Orlando, Florida.
Kolata, G. 1985. How safe are engineered organisms? Science 229:34–35.
Barry, G.F. 1986. The permanent insertion of foreign genes into the chromosome of soil bacteria. Bio/Technology (May).
Matteucci, M.D. and Caruthers, M.H. 1981. Synthesis of deoxyolionucleotides on a polymer support. J. Am. Chem. Soc. 103:3185–3191.
Guerry, P., Embden, J.V., and Falkow, S. 1974. Molecular nature of two nonconjugative plasmids carrying drug resistance genes. J. Bacteriol. 117:619–630.
Messing, F., Crea, R., and Seeburg, P.H. 1981. A system for shotgun DNA sequencing. Nucleic Acids Res. 9:309–318.
King, E.O., Ward, M.K., and Raney, D.E. 1954. Two simple media for the demonstration of pycocyanin and fluorescein. J. Lab. Clin. Med. 44:301–307.
Casadaban, M. and Cohen, S. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179–207.
Poindron, P., Bourguignat, A., Scheid, F., and Lombard, Y. 1977. The orthonitrophenyl-β-D-galactoside hydrolase from Pseudomonas aeruginosa (0:11 serotypes) is a superficial enzyme. Can. J. Microbiol. 23:798–810.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Drahos, D., Hemming, B. & McPherson, S. Tracking Recombinant Organisms in the Environment: β–Galactosidase as a Selectable Non–Antibiotic Marker for Fluorescent Pseudomonads. Nat Biotechnol 4, 439–444 (1986). https://doi.org/10.1038/nbt0586-439
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nbt0586-439