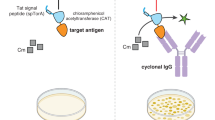
Abstract
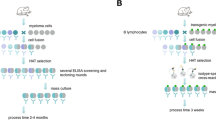
We present a simple and flexible procedure to elicit and assay anti-peptide antibodies without peptide synthesis. It consists of expressing the peptide of interest in the form of a genetic insert within two different “recipient” bacterial proteins. One hybrid protein is used as immunogen for the induction of antibodies against the inserted peptide and the other as antigen for monitoring the anti-peptide antibodies raised. The two “recipient” proteins used are the MalE and the LamB proteins from E. coli. The MalE hybrid proteins can be affinity purified on an amylose column using mild nondenaturing conditions and can be crystalized for structural studies; LamB hybrid proteins express the inserted peptide on the cell surface so that intact bacteria can be used as a reagent. We chose, as a model peptide, a B-cell epitope from the pre-S(2) region of Hepatitis B virus. With both MalE and LamB hybrid proteins, high titres of anti-preS antibodies, able to react with native HBsAg particles, were induced in mice. The anti-peptide antibody titres recorded by ELISA were comparable to those obtained when either a synthetic peptide, or the hybrid proteins, were used as immobilized antigen.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
van Regenmortel, M.H.V. 1989. Structural and functional approaches to the study of protein antigenicity. Immunol. Today 10: 266–271.
Lerner, R.A. 1984. Antibodies of predetermined specificity in biology and medicine. Adv. Immunol. 36: 1–44.
Arnon, R. 1986. Synthetic peptides as the basis for vaccines. Trends in Biochem. Sci. 11: 521–524.
Leclerc, C., Przewlocki, G., Shultze, M.P. and Chedid, L. 1987. A synthetic vaccine constructed by copolymerization of B and T cell determinants. Eur. J. Immunol. 17: 269–273.
Charbit, A. 1990. Topologie fonctionnelle de LamB, une protéine de la membrane externe d'E. coli K12. Implications pour la mise au point de vaccins bactériens. University Thesis, University Paris VII, Paris, France.
Charbit, A., Boulain, J.-C., Ryter, A. and Hofnung, M. 1986. Probing the topology of a bacterial membrane protein by genetic insertion of a foreign epitope, Expression at the cell surface. EMBO J. 5: 3029–3037.
Agterberg, M., Adriaanse, H. and Tommassen, J. 1987. Use of outer membrane protein PhoE as a carrier for the transport of a foreign anigenic determinant to the cell surface of Escherichia coli K12. Gene 59: 145–150.
Scott, J.K. and Smith, G.P. 1990. Searching for peptide ligands with an epitope library. Science 249: 386–390.
Hofnung, M., Bedouelle, H., Boulain, J.-C., Clément, J.-M., Charbit, A., Duplay, P., Gehring, K., Martineau, P., Saurin, W. and Szmelcman, S. 1988. Genetic approaches to the study and the use of proteins: random point mutations and random linker insertions. Bull. Inst. Pasteur 86: 95–101.
Charbit, A., Leclerc, C., van der Werf, S., Martineau, P., Ronco, J., O'Callaghan, D. and Hofnung, M. 1990. Antibody response to foreign epitopes expressed within envelope proteins of Gram− bacteria, p. 451–455. In: Vaccines 90. F. Brown, R. Chanock, H. Ginsberg and R. Lerner (Eds.). Cold Spring Harbor Laboratory, NY.
Leclerc, C., Martineau, P., Van der Werf, S., Deriaud, E., Duplay, P. and Hofnung, M. 1990. Induction of virus-neutralizing antibodies by bacteria expressing the C3 poliovirus epitope in the periplasm. J. Immunology 14: 3174–3182.
Charbit, A., Sobczak, E., Michel, M.L., Molla, A., Tiollais, P. and Hofnung, M. 1987. Presentation of two epitopes of the preS2 region of Hepatitis B on live recombinant bacteria. J. of Immunology 139: 1658–1664.
Leclerc, C., Charbit, A., Molla, A. and Hofnung, M. 1989. Antibody response to a foreign epitope expressed at the surface of recombinant bacteria: importance of the route of immunization. Vaccine 7: 242– 248.
Charbit, A., van der Werf, S., Boulain, J.-C., Girard, M. and Hofnung, M. 1988. Expression of a poliovirus neutralization epitope at the surface of recombinant bacteria: First immunization results. Ann. Microbiol. (Institut Pasteur) 139: 45–58.
Ferenci, T. and Klotz, U. 1978. Affinity chromatographic isolation of the periplasmic maltose binding protein of Escherichia coli. FEBS Letts. 94: 213–217.
Millich, D.R., McLachlan, A., Chisari, F.V. and Thornton, G.B. 1986. Nonverlapping T and B cell determinants on an hepatitis B surface antigen Pre-S(2) region synthetic peptide. J. Exp. Med. 164: 532–547.
Rodseth, L.E., Martineau, P., Duplay, P., Hofnung, M. and Quiocho, F.A. 1990. Crystallization of genetically engineered active maltose-binding proteins, including an immunogenic viral epitope insertion. J. Mol. Biol. 213: 607–611.
Spurlino, J.C. 1988. The 3-D structure of D-maltose binding protein from E. coli. PhD Thesis, Rice University, Houston Tx., USA.
van der Werf, S., Charbit, A., Leclerc, C., Mimic, V., Ronco, J., Girard, M. and Hofnung, M. 1990. Critical role of neighbouring sequences on the immunogenicity of the C3 poliovirus neutralization epitope expressed at the surface of recombinant bacteria. Vaccine 8: 269–277.
Stahi, S., Sjolander, A., Nygren, P.-A., Berzins, K., Perlmann, P. and Ulhén, M. 1989. A dual expression system for the generation, analysis and purification of antibodies to a repeated sequence of the Plasmodium falciparum antigen Pf155/RESA. J. of Immunol. Meth. 124: 43–52.
Charbit, A., Molla, A., Saurin, W. and Hofnung, M. 1988. Versatility of a vector for expressing foreign polypeptides at the surface of Gram− bacteria. Gene 70: 181–189.
Miller, J.H. 1972. Experiments in Molecular Genetics, p. 431. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York.
Bouges-Bocquet, B., Villarroya, H. and Hofnung, M. 1984. Linker mutagenesis in the gene of an outer membrane protein of E. coli. J. Cell. Biochem. 24: 217–228.
van Regenmortel, M.H.V., Briand, J.P., Muller, S. and Plaué, S. 1988. Synthetic Polypeptides as Antigens, p. 131–144. Elsevier, Amsterdam, NY and Oxford.
Neu, H.C., Heppel, L.A. 1965. The release of enzymes of E. coli by osmotic shock during the formation of spheroplasts. J. Biol. Chem. 240: 3685–3692.
Sambrook, J., Fritsch, E.F. and Maniatis, T. 1989. Molecular Cloning, a Laboratory Manual, 2nd Edition, Volume III, p. 18.47–18.75.
Molla, A., Charbit, A., Le Guern, A., Ryter, A. and Hofnung, M., 1989. Antibodies against synthetic peptides and the topology of LamB, an outer membrane protein from Escherichia coli K12. Biochemistry 28: 8934–8941.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Martineau, P., Charbit, A., Leclerc, C. et al. A Genetic System to Elicit and Monitor Anti-Peptide Antibodies Without Peptide Synthesis. Nat Biotechnol 9, 170–172 (1991). https://doi.org/10.1038/nbt0291-170
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nbt0291-170
This article is cited by
-
Surface display of recombinant proteins on Escherichia coli by BclA exosporium of Bacillus anthracis
Microbial Cell Factories (2013)
-
Investigation of spore coat display of Bacillus subtilis β-galactosidase for developing of whole cell biocatalyst
Archives of Microbiology (2013)
-
Surface display of recombinant protein on the cell surface of Bacillus subtilis by the CotB anchor protein
World Journal of Microbiology and Biotechnology (2011)