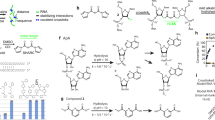
Abstract
Gene transcription is regulated by proteins that bind specific DNA sequences and control the initiation of RNA synthesis. A major challenge is to map all of the regulatory sites in the genome and to identify the proteins that bind them. Because members of transcription factor families often exhibit similar sequence preferences, methods for determining intermolecular contacts in protein–DNA interfaces must be sensitive to even subtle structural differences. The most detailed structural views of protein–DNA interfaces have been obtained through X-ray crystallography and NMR spectroscopy, and these methods have revolutionized the understanding of the structural determinants of sequence-specific recognition1. Neither crystallography nor NMR, however, is particularly well-suited to high-throughput applications such as pan-genomic elucidation of regulatory sequences; in addition, these methods yield no information on the energetic contribution of particular contacts. Here we report a straightforward, high-resolution biochemical method for mapping, at single-nucleotide resolution, DNA bases that are subject to sequence-specific contacts by regulatory proteins.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Pabo, C.O. & Nekludova, L. Geometric analysis and comparison of protein–DNA interfaces: why is there no simple code for recognition? J. Mol. Biol. 301, 597–624 (2000).
Siebenlist, U. & Gilbert, W. Contacts between Escherichia coli RNA polymerase and an early promotor of phage T7. Proc. Natl. Acad. Sci. USA 77, 122–126 (1980).
Hayes, J.J. & Tullius, T.D. The missing nucleoside experiment: a new technique to study recognition of DNA by protein. Biochemistry 28, 9521–9527 (1989).
Brunelle, A. & Schleif, R.F. Missing contact probing of DNA–protein interactions. Proc. Natl. Acad. Sci. USA 84, 6673–6676 (1987).
Kalnik, M.W., Chang, C., Johnson, F., Grollman, A.P. & Patel, D.J. NMR studies of abasic sites in DNA duplexes: deoxyadenosine stacks into the helix opposite acyclic lesions. Biochemistry 28, 3373–3383 (1989).
Jordan, S.R. & Pabo, C.O. Structure of the lambda complex at 2.5 Å resolution: details of the repressor–operator interactions. Science 242, 893–899 (1988).
Hayashibara, K.C. & Verdine, G.L. Template-directed interference footprinting of protein–guanine contacts in DNA. J. Am. Chem. Soc. 113, 5104–5106 (1991).
Hayashibara, K.C. & Verdine, G.L. Template-directed interference footprinting of cytosine contacts in a protein-DNA complex: potent interference by 5-aza-2'-deoxycytidine. Biochemistry 31, 11265–11273 (1992).
Mascarenas, J.L., Hayashibara, K.C. & Verdine, G.L. Template-directed interference footprinting of protein-thymine contacts. J. Am. Chem. Soc. 115, 373–374 (1993).
Min, C., Cushing, T.D. & Verdine, G.L. Template-directed interference footprinting of protein–adenine contacts. J. Am. Chem. Soc. 118, 6116–6120 (1996).
Landini, P. & Volkert, M.R. Transcriptional activation of the Escherichia coli adaptive response gene aidB is mediated by binding of methylated Ada protein. Evidence for a new consensus sequence for Ada-binding sites. J. Biol. Chem. 270, 8285–8289 (1995).
Sedgwick, B., Robins, P., Totty, N. & Lindahl, T. Functional domains and methyl acceptor sites of the Escherichia coli ada protein. J. Biol. Chem. 263, 4430–4433 (1988).
Landini, P., Bown, J.A., Volkert, M.R. & Busby, S.J.W. Ada protein–RNA polymerase σ subunit interaction and α-subunit–promoter DNA interaction are necessary at different steps in transcription initiation at the Escherichia coli ada and aidB promoters. J. Biol. Chem. 273, 13307–13312 (1998).
Maxam, A.M. & Gilbert, W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 65, 499–560 (1980).
Teo, I., Sedgwick, B., Kilpatrick, M. W., McCarthy, T. V. & Lindahl, T. The intracellular signal for induction of resistance to alkylating agents in E. coli. Cell 45, 315–324 (1986).
Ross, W., Ernst, A. & Gourse, R.L. Fine structure of E. coli RNA polymerase–promoter interactions: α subunit binding to the UP element minor groove. Genes Dev. 15, 491–506 (2001).
Lander, E.S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Venter, J.C. et al. The sequence of the human genome. Science 291, 1304–1351 (2001).
Lockhart, D.J. & Winzeler, E.A. Genomics, gene expression and DNA arrays. Nature 405, 827–836 (2000).
Kovacs, T. & Otvos, L. Simple synthesis of 5-vinyl- and 5-ethynyl-2'-deoxyuridine-5′-triphosphates. Tetrahedron Lett. 29, 4525–4528 (1988).
Acknowledgements
This work was supported by a grant from Variagenics, Inc. M.J.S. is the recipient of fellowships from the Alfred and Isabel Bader Foundation and from Hoffman-La Roche and Eli Lilly. A.E. gratefully acknowledges support from the Swiss National Science Foundation (postdoctoral fellowship 1998–1999) and from Harvard University (1999–2000). We thank Jia Wolfe, Jeff Olson, and Tomohiko Kawate of Variagenics, Inc. for sharing unpublished results, and members of the Verdine group for valuable discussions.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Rights and permissions
About this article
Cite this article
Storek, M., Ernst, A. & Verdine, G. High-resolution footprinting of sequence-specific protein–DNA contacts. Nat Biotechnol 20, 183–186 (2002). https://doi.org/10.1038/nbt0202-183
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nbt0202-183
This article is cited by
-
Detection of plasmid DNA vectors following gene transfer to the murine airways
Gene Therapy (2005)
-
Binding assays get into the groove
Nature Biotechnology (2002)