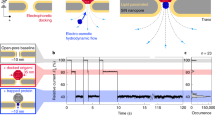
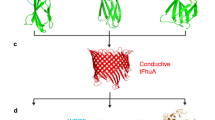
Abstract
Protein–protein interactions (PPIs) are essential for many cellular processes. However, transient PPIs are difficult to measure at high throughput or in complex biological fluids using existing methods. We engineered a genetically encoded sensor for real-time sampling of transient PPIs at single-molecule resolution. Our sensor comprises a truncated outer membrane protein pore, a flexible tether, a protein receptor and a peptide adaptor. When a protein ligand present in solution binds to the receptor, reversible capture and release events of the receptor can be measured as current transitions between two open substates of the pore. Notably, the binding and release of the receptor by a protein ligand can be unambiguously discriminated in a complex sample containing fetal bovine serum. Our selective nanopore sensor could be applied for single-molecule protein detection, could form the basis for a nanoproteomics platform or might be adapted to build tools for protein profiling and biomarker discovery.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hayes, S., Malacrida, B., Kiely, M. & Kiely, P.A. Studying protein-protein interactions: progress, pitfalls and solutions. Biochem. Soc. Trans. 44, 994–1004 (2016).
Yoo, J., Lee, T.S., Choi, B., Shon, M.J. & Yoon, T.Y. Observing extremely weak protein-protein interactions with conventional single-molecule fluorescence microscopy. J. Am. Chem. Soc. 138, 14238–14241 (2016).
De Keersmaecker, H. et al. Mapping transient protein interactions at the nanoscale in living mammalian cells. ACS Nano 12, 9842–9854 (2018).
Nogal, B., Bowman, C.A. & Ward, A.B. Time-course, negative-stain electron microscopy-based analysis for investigating protein-protein interactions at the single-molecule level. J. Biol. Chem. 292, 19400–19410 (2017).
Gonzalez, L.C. Protein microarrays, biosensors, and cell-based methods for secretome-wide extracellular protein-protein interaction mapping. Methods 57, 448–458 (2012).
Douzi, B. Protein-protein interactions: surface plasmon resonance. Methods Mol. Biol. 1615, 257–275 (2017).
Pierce, M.M., Raman, C.S. & Nall, B.T. Isothermal titration calorimetry of protein-protein interactions. Methods 19, 213–221 (1999).
Sackmann, B. & Neher, E. Single-Channel Recording. Second edn. (Kluwer Academic/Plenum, New York, 1995).
Wei, R., Gatterdam, V., Wieneke, R., Tampé, R. & Rant, U. Stochastic sensing of proteins with receptor-modified solid-state nanopores. Nat. Nanotechnol. 7, 257–263 (2012).
Ying, Y.L., Yu, R.J., Hu, Y.X., Gao, R. & Long, Y.T. Single antibody-antigen interactions monitored via transient ionic current recording using nanopore sensors. Chem. Commun. (Camb.) 53, 8620–8623 (2017).
Weichbrodt, C. et al. Antibiotic translocation through porins studied in planar lipid bilayers using parallel platforms. Analyst 140, 4874–4881 (2015).
Reiner, J.E. et al. Disease detection and management via single nanopore-based sensors. Chem. Rev. 112, 6431–6451 (2012).
Deamer, D., Akeson, M. & Branton, D. Three decades of nanopore sequencing. Nat. Biotechnol. 34, 518–524 (2016).
Ayub, M. & Bayley, H. Engineered transmembrane pores. Curr. Opin. Chem. Biol. 34, 117–126 (2016).
Burns, J.R., Seifert, A., Fertig, N. & Howorka, S. A biomimetic DNA-based channel for the ligand-controlled transport of charged molecular cargo across a biological membrane. Nat. Nanotechnol. 11, 152–156 (2016).
Howorka, S. Building membrane nanopores. Nat. Nanotechnol. 12, 619–630 (2017).
Movileanu, L., Howorka, S., Braha, O. & Bayley, H. Detecting protein analytes that modulate transmembrane movement of a polymer chain within a single protein pore. Nat. Biotechnol. 18, 1091–1095 (2000).
Rotem, D., Jayasinghe, L., Salichou, M. & Bayley, H. Protein detection by nanopores equipped with aptamers. J. Am. Chem. Soc. 134, 2781–2787 (2012).
Harrington, L., Cheley, S., Alexander, L.T., Knapp, S. & Bayley, H. Stochastic detection of Pim protein kinases reveals electrostatically enhanced association of a peptide substrate. Proc. Natl. Acad. Sci. USA 110, E4417–E4426 (2013).
Thakur, A.K., Larimi, M.G., Gooden, K. & Movileanu, L. Aberrantly large single-channel conductance of polyhistidine arm-containing protein nanopores. Biochemistry 56, 4895–4905 (2017).
Locher, K.P. et al. Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes. Cell 95, 771–778 (1998).
Schreiber, G. & Fersht, A.R. Interaction of barnase with its polypeptide inhibitor barstar studied by protein engineering. Biochemistry 32, 5145–5150 (1993).
Schreiber, G. & Fersht, A.R. Energetics of protein-protein interactions: analysis of the barnase-barstar interface by single mutations and double mutant cycles. J. Mol. Biol. 248, 478–486 (1995).
Deyev, S.M., Waibel, R., Lebedenko, E.N., Schubiger, A.P. & Plückthun, A. Design of multivalent complexes using the barnase*barstar module. Nat. Biotechnol. 21, 1486–1492 (2003).
Kudlinzki, D., Schmitt, A., Christian, H. & Ficner, R. Structural analysis of the C-terminal domain of the spliceosomal helicase Prp22. Biol. Chem. 393, 1131–1140 (2012).
Mohammad, M.M., Howard, K.R. & Movileanu, L. Redesign of a plugged beta-barrel membrane protein. J. Biol. Chem. 286, 8000–8013 (2011).
Mohammad, M.M. et al. Engineering a rigid protein tunnel for biomolecular detection. J. Am. Chem. Soc. 134, 9521–9531 (2012).
Perkins, J.R., Diboun, I., Dessailly, B.H., Lees, J.G. & Orengo, C. Transient protein-protein interactions: structural, functional, and network properties. Structure 18, 1233–1243 (2010).
Nivala, J., Mulroney, L., Li, G., Schreiber, J. & Akeson, M. Discrimination among protein variants using an unfoldase-coupled nanopore. ACS Nano 8, 12365–12375 (2014).
Kennedy, E., Dong, Z., Tennant, C. & Timp, G. Reading the primary structure of a protein with 0.07 nm3 resolution using a subnanometre-diameter pore. Nat. Nanotechnol. 11, 968–976 (2016).
Sze, J.Y.Y., Ivanov, A.P., Cass, A.E.G. & Edel, J.B. Single molecule multiplexed nanopore protein screening in human serum using aptamer modified DNA carriers. Nat. Commun. 8, 1552 (2017).
Huang, G., Willems, K., Soskine, M., Wloka, C. & Maglia, G. Electro-osmotic capture and ionic discrimination of peptide and protein biomarkers with FraC nanopores. Nat. Commun. 8, 935 (2017).
Restrepo-Pérez, L., Joo, C. & Dekker, C. Paving the way to single-molecule protein sequencing. Nat. Nanotechnol. 13, 786–796 (2018).
Buckle, A.M., Schreiber, G. & Fersht, A.R. Protein-protein recognition: crystal structural analysis of a barnase-barstar complex at 2.0-A resolution. Biochemistry 33, 8878–8889 (1994).
Guillet, V., Lapthorn, A., Hartley, R.W. & Mauguen, Y. Recognition between a bacterial ribonuclease, barnase, and its natural inhibitor, barstar. Structure 1, 165–176 (1993).
McManus, O.B. & Magleby, K.L. Kinetic states and modes of single large-conductance calcium-activated potassium channels in cultured rat skeletal muscle. J. Physiol. (Lond.) 402, 79–120 (1988).
Acknowledgements
We thank S. Loh for generosity in using his FPLC instrument in the very early stages of these studies and A. Matouschek (University of Texas at Austin) for his kindness in offering plasmids containing genes that encode Bn and Bs proteins, as well as M.L. Ghahari and M.M. Mohammad for their assistance in the very early stages of this project. This work was supported by US National Institutes of Health grants GM088403 (L.M.) and GM129429 (L.M.).
Author information
Authors and Affiliations
Contributions
A.K.T. and L.M. designed research. A.K.T. performed research and analyzed data. A.K.T. and L.M. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
A.K.T. and L.M. are named inventors on two provisional patent applications (US 62/720,190 and US 62/579,982) filed by Syracuse University on this work.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–14 and Supplementary Tables 1–8 (PDF 6811 kb)
Rights and permissions
About this article
Cite this article
Thakur, A., Movileanu, L. Real-time measurement of protein–protein interactions at single-molecule resolution using a biological nanopore. Nat Biotechnol 37, 96–101 (2019). https://doi.org/10.1038/nbt.4316
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.4316
This article is cited by
-
Nanopore DNA sequencing technologies and their applications towards single-molecule proteomics
Nature Chemistry (2024)
-
Single-molecule fingerprinting of protein-drug interaction using a funneled biological nanopore
Nature Communications (2023)
-
A generalizable nanopore sensor for highly specific protein detection at single-molecule precision
Nature Communications (2023)
-
Single-exonuclease nanocircuits reveal the RNA degradation dynamics of PNPase and demonstrate potential for RNA sequencing
Nature Communications (2023)
-
Analytical device miniaturization for the detection of circulating biomarkers
Nature Reviews Bioengineering (2023)