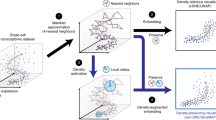
Abstract
Advances in single-cell technologies have enabled high-resolution dissection of tissue composition. Several tools for dimensionality reduction are available to analyze the large number of parameters generated in single-cell studies. Recently, a nonlinear dimensionality-reduction technique, uniform manifold approximation and projection (UMAP), was developed for the analysis of any type of high-dimensional data. Here we apply it to biological data, using three well-characterized mass cytometry and single-cell RNA sequencing datasets. Comparing the performance of UMAP with five other tools, we find that UMAP provides the fastest run times, highest reproducibility and the most meaningful organization of cell clusters. The work highlights the use of UMAP for improved visualization and interpretation of single-cell data.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Saeys, Y., Van Gassen, S. & Lambrecht, B.N. Computational flow cytometry: helping to make sense of high-dimensional immunology data. Nat. Rev. Immunol. 16, 449–462 (2016).
Tenenbaum, J.B., De Silva, V. & Langford, J.C. A global geometric framework for nonlinear dimensionality reduction. Science 290, 2319–2323 (2000).
Coifman, R.R. et al. Geometric diffusions as a tool for harmonic analysis and structure definition of data: diffusion maps. Proc. Natl. Acad. Sci. USA 102, 7426–7431 (2005).
Van Der Maaten, L. & Hinton, G. Visualizing high-dimensional data using t-SNE. journal of machine learning research. J. Mach. Learn. Res. 9, 26 (2008).
Amir, A.D. et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol. 31, 545–552 (2013).
van Unen, V. et al. Mass cytometry of the human mucosal immune system identifies tissue- and disease-associated immune subsets. Immunity 44, 1227–1239 (2016).
McInnes, L. & Healy, J. UMAP: uniform manifold approximation and projection for dimension reduction. Preprint at https://arxiv.org/abs/1802.03426 (2018).
McInnes, L., Healy, J., Saul, N. & Großberger, L. UMAP: uniform manifold approximation and projection. J. Open Source Softw. 3, 861 (2018).
Han, X. et al. Mapping the mouse cell atlas by microwell-seq. Cell 172, 1091–1107.e17 (2018).
Samusik, N., Good, Z., Spitzer, M.H., Davis, K.L. & Nolan, G.P. Automated mapping of phenotype space with single-cell data. Nat. Methods 13, 493–496 (2016).
Wong, M.T. et al. A high-dimensional atlas of human T cell diversity reveals tissue-specific trafficking and cytokine signatures. Immunity 45, 442–456 (2016).
Van Der Maaten, L. Accelerating t-SNE using tree-based algorithms. J. Mach. Learn. Res. 15, 3221–3245 (2014).
Linderman, G.C., Rachh, M., Hoskins, J.G., Steinerberger, S. & Kluger, Y. Efficient algorithms for t-distributed stochastic neighborhood embedding. Preprint at https://arxiv.org/abs/1712.09005 (2017).
Ding, J., Condon, A. & Shah, S.P. Interpretable dimensionality reduction of single cell transcriptome data with deep generative models. Nat. Commun. 9, 2002 (2018).
Levine, J.H. et al. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell 162, 184–197 (2015).
Huang, H., Li, Y. & Liu, B. Transcriptional regulation of mast cell and basophil lineage commitment. Semin. Immunopathol. 38, 539–548 (2016).
Wattenberg, M., Viégas, F. & Johnson, I. How to use t-SNE effectively. Distill 1, e2 (2016).
de Graaf, C.A. et al. Haemopedia: an expression atlas of murine hematopoietic cells. Stem Cell Rep. 7, 571–582 (2016).
Mårtensson, I.-L., Keenan, R.A. & Licence, S. The pre-B-cell receptor. Curr. Opin. Immunol. 19, 137–142 (2007).
Wolf, F.A., Angerer, P. & Theis, F.J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Aibar, S. et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086 (2017).
Acknowledgements
We thank members of the Singapore Immunology Network and notably members of the E.W.N. laboratory. We thank S. Li, Y. Simoni, M. Chng, Y. Cheng, J.W. Lim and M. Fehlings for their insightful feedback. This study was funded by A-STAR/SIgN core funding and A-STAR/SIgN immunomonitoring platform funding.
Author information
Authors and Affiliations
Contributions
E.B., L.M., J.H., C.-A.D., I.W.H.K. and E.W.N. analyzed data. L.G.N., F.G. and E.W.N. helped supervise the project. L.M. and J.H. developed UMAP. All authors participated in writing and revising the manuscript.
Corresponding author
Ethics declarations
Competing interests
E.W.N. is a board director and shareholder of immunoSCAPE Pte. Ltd., which is an immune profiling service provider.
Integrated supplementary information
Supplementary Figure 1 Phenograph clustering identifies cell clusters in the Wong dataset
a) Phenotypic characterization of the phenograph clusters. Each cluster medoid is represented after column-wise Z-score transformation. b) Identification of each phenograph cluster of both UMAP (left), t-SNE (middle) and 2D PCA (right). For clarity, only twelve clusters are shown per plot.
Supplementary Figure 2 Annotation of the tissue of origins on UMAP, t-SNE and PCA plots
Scatterplot of embeddings of the Wong dataset using UMAP (top), t-SNE (middle) and 2D PCA (bottom) color-coded by tissues of origin.
Supplementary Figure 3 Identification of unlabeled erythrocytes in the Samusik_01 dataset
Expression of Ter119 (a marker for mature erythrocytes) color-coded on the UMAP embedding of the Samusik_01 dataset.
Supplementary Figure 4 Surface densities of events in UMAP and t-SNE embeddings
Heatmap of the density of a 300x300 square grid of the UMAP or t-SNE projections for the Samusik_01 dataset. The number of events in each bin is color-coded.
Supplementary Figure 5 Pre-filtering of the Han dataset
Top: UMAP projection of the full Han dataset annotated by AUC scores for various cell lineages (red: high score, blue: low score). Bottom: full Han dataset colored by sample type, Sample ID and pre-filtering status.
Supplementary Figure 6 Side-by-side comparison of each dimensionality reduction method across all datasets annotated by cell types.
Scatterplots of six dimensionality-reduction methods and 6 datasets. Cell populations are annotated using manual gating (Samusik dataset), manually-labelled Phenograph clusters (Wong dataset) or sample of origin (Han_400k dataset).
Supplementary Figure 7 Qualitative assessment of the reproducibility of embeddings
Embeddings of full datasets as well as subsamples of varying sizes replicated thrice for five dimensionality reduction methods. The color-code is generated using the embedding of the full dataset and propagated to the subsamples. Datasets shown are the a) Samusik_all, b) Wong and c) Han_400k datasets.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 (PDF 1451 kb)
Supplementary Table 1
Description of the datasets (XLSX 5 kb)
Supplementary Table 2
Algorithms benchmarked (XLSX 5 kb)
Rights and permissions
About this article
Cite this article
Becht, E., McInnes, L., Healy, J. et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol 37, 38–44 (2019). https://doi.org/10.1038/nbt.4314
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.4314
This article is cited by
-
Sex disparities revealed by single-cell and bulk sequencing and their impacts on the efficacy of immunotherapy in esophageal cancer
Biology of Sex Differences (2024)
-
Systematic immune cell dysregulation and molecular subtypes revealed by single-cell RNA-seq of subjects with type 1 diabetes
Genome Medicine (2024)
-
Differentiation is accompanied by a progressive loss in transcriptional memory
BMC Biology (2024)
-
Strategies for improving the performance of prediction models for response to immune checkpoint blockade therapy in cancer
BMC Research Notes (2024)
-
Benchmarking differential abundance methods for finding condition-specific prototypical cells in multi-sample single-cell datasets
Genome Biology (2024)