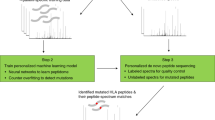
Abstract
Neoantigens, which are expressed on tumor cells, are one of the main targets of an effective antitumor T-cell response. Cancer immunotherapies to target neoantigens are of growing interest and are in early human trials, but methods to identify neoantigens either require invasive or difficult-to-obtain clinical specimens, require the screening of hundreds to thousands of synthetic peptides or tandem minigenes, or are only relevant to specific human leukocyte antigen (HLA) alleles. We apply deep learning to a large (N = 74 patients) HLA peptide and genomic dataset from various human tumors to create a computational model of antigen presentation for neoantigen prediction. We show that our model, named EDGE, increases the positive predictive value of HLA antigen prediction by up to ninefold. We apply EDGE to enable identification of neoantigens and neoantigen-reactive T cells using routine clinical specimens and small numbers of synthetic peptides for most common HLA alleles. EDGE could enable an improved ability to develop neoantigen-targeted immunotherapies for cancer patients.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
18 December 2018
Supplementary Data 6 as originally posted was actually Supplementary Data 5, Supplementary Data 7 as originally posted was actually Supplementary Data 6, Supplementary Data 8 as originally posted was actually Supplementary Data 7, Supplementary Data 9 as originally posted was actually Supplementary Data 8, and Supplementary Data 5 as originally posted was actually a corrupted version of Supplementary Data 9. The error has been corrected online as of 18 December 2018.
References
Snyder, A. et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 371, 2189–2199 (2014).
Rizvi, N.A. et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348, 124–128 (2015).
Carreno, B.M. et al. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science 348, 803–808 (2015).
Ott, P.A. et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 547, 217–221 (2017).
Sahin, U. et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 547, 222–226 (2017).
Tran, E. et al. T-cell transfer therapy targeting mutant KRAS in cancer. N. Engl. J. Med. 375, 2255–2262 (2016).
Gros, A. et al. Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat. Med. 22, 433–438 (2016).
Anonymous. The problem with neoantigen prediction. Nat. Biotechnol. 35, 97 (2017).
Vitiello, A. & Zanetti, M. Neoantigen prediction and the need for validation. Nat. Biotechnol. 35, 815–817 (2017).
Bassani-Sternberg, M., Pletscher-Frankild, S., Jensen, L.J. & Mann, M. Mass spectrometry of human leukocyte antigen class I peptidomes reveals strong effects of protein abundance and turnover on antigen presentation. Mol. Cell. Proteomics 14, 658–673 (2015).
Vita, R. et al. The Immune Epitope Database (IEDB) 3.0. Nucleic Acids Res. 43, D405–D412 (2015).
Andreatta, M. & Nielsen, M. Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics 32, 511–517 (2016).
O'Donnell, T.J. et al. MHCflurry: open-source class I MHC binding affinity prediction. Cell Syst. 7, 129–132.e4 (2018).
Bassani-Sternberg, M. et al. Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat. Commun. 7, 13404 (2016).
Abelin, J.G. et al. Mass Spectrometry profiling of HLA-associated peptidomes in mono-allelic cells enables more accurate epitope prediction. Immunity 46, 315–326 (2017).
Yadav, M. et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature 515, 572–576 (2014).
Stranzl, T., Larsen, M.V., Lundegaard, C. & Nielsen, M. NetCTLpan: pan-specific MHC class I pathway epitope predictions. Immunogenetics 62, 357–368 (2010).
Jurtz, V. et al. NetMHCpan-4.0: improved peptide-MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J. Immunol. 199, 3360–3368 (2017).
Bentzen, A.K. et al. Large-scale detection of antigen-specific T cells using peptide-MHC-I multimers labeled with DNA barcodes. Nat. Biotechnol. 34, 1037–1045 (2016).
Tran, E. et al. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science 350, 1387–1390 (2015).
Strønen, E. et al. Targeting of cancer neoantigens with donor-derived T cell receptor repertoires. Science 352, 1337–1341 (2016).
Trolle, T. et al. The length distribution of class I-restricted T cell epitopes is determined by both peptide supply and MHC allele-specific binding preference. J. Immunol. 196, 1480–1487 (2016).
Di Marco, M. et al. Unveiling the peptide motifs of HLA-C and HLA-G from naturally presented peptides and generation of binding prediction matrices. J. Immunol. 199, 2639–2651 (2017).
Goodfellow, I., Bengio, Y. & Courville, A. Deep Learning (MIT Press, 2016).
Sette, A. et al. The relationship between class I binding affinity and immunogenicity of potential cytotoxic T cell epitopes. J. Immunol. 153, 5586–5592 (1994).
Fortier, M.-H. et al. The MHC class I peptide repertoire is molded by the transcriptome. J. Exp. Med. 205, 595–610 (2008).
Pearson, H. et al. MHC class I-associated peptides derive from selective regions of the human genome. J. Clin. Invest. 126, 4690–4701 (2016).
Bassani-Sternberg, M. et al. Deciphering HLA-I motifs across HLA peptidomes improves neo-antigen predictions and identifies allostery regulating HLA specificity. PLoS Comput. Biol. 13, e1005725 (2017).
Andreatta, M., Lund, O. & Nielsen, M. Simultaneous alignment and clustering of peptide data using a Gibbs sampling approach. Bioinformatics 29, 8–14 (2013).
Andreatta, M., Alvarez, B. & Nielsen, M. GibbsCluster: unsupervised clustering and alignment of peptide sequences. Nucleic Acids Res. 45, W458–W463 (2017).
Zacharakis, N. et al. Immune recognition of somatic mutations leading to complete durable regression in metastatic breast cancer. Nat. Med. 24, 724–730 (2018).
Chudley, L. et al. Harmonisation of short-term in vitro culture for the expansion of antigen-specific CD8+ T cells with detection by ELISPOT and HLA-multimer staining. Cancer Immunol. Immunother. 63, 1199–1211 (2014).
Van Allen, E.M. et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 350, 207–211 (2015).
Anagnostou, V. et al. Evolution of neoantigen landscape during immune checkpoint blockade in non-small cell lung cancer. Cancer Discov. 7, 264–276 (2017).
Stevanović, S. et al. Landscape of immunogenic tumor antigens in successful immunotherapy of virally induced epithelial cancer. Science 356, 200–205 (2017).
Gillette, M.A. & Carr, S.A. Quantitative analysis of peptides and proteins in biomedicine by targeted mass spectrometry. Nat. Methods 10, 28–34 (2013).
Boegel, S., Löwer, M., Bukur, T., Sahin, U. & Castle, J.C. A catalog of HLA type, HLA expression, and neo-epitope candidates in human cancer cell lines. Oncoimmunology 3, e954893 (2014).
Johnson, D.B. et al. Melanoma-specific MHC-II expression represents a tumour-autonomous phenotype and predicts response to anti-PD-1/PD-L1 therapy. Nat. Commun. 7, 10582 (2016).
Robbins, P.F. et al. A pilot trial using lymphocytes genetically engineered with an NY-ESO-1-reactive T-cell receptor: long-term follow-up and correlates with response. Clin. Cancer Res. 21, 1019–1027 (2015).
Calis, J.J.A. et al. Properties of MHC class I presented peptides that enhance immunogenicity. PLoS Comput. Biol. 9, e1003266 (2013).
Duan, F. et al. Genomic and bioinformatic profiling of mutational neoepitopes reveals new rules to predict anticancer immunogenicity. J. Exp. Med. 211, 2231–2248 (2014).
Glanville, J. et al. Identifying specificity groups in the T cell receptor repertoire. Nature 547, 94–98 (2017).
Dash, P. et al. Quantifiable predictive features define epitope-specific T cell receptor repertoires. Nature 547, 89–93 (2017).
Hunt, D.F. et al. Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science 255, 1261–1263 (1992).
Zarling, A.L. et al. Identification of class I MHC-associated phosphopeptides as targets for cancer immunotherapy. Proc. Natl. Acad. Sci. USA 103, 14889–14894 (2006).
Eng, J.K., Jahan, T.A. & Hoopmann, M.R. Comet: an open-source MS/MS sequence database search tool. Proteomics 13, 22–24 (2013).
Käll, L., Storey, J.D., MacCoss, M.J. & Noble, W.S. Assigning significance to peptides identified by tandem mass spectrometry using decoy databases. J. Proteome Res. 7, 29–34 (2008).
Li, B. & Dewey, C.N. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011).
Chollet, F. et al. Keras https://keras.io (2015).
Al-Rfou, R. et al. Theano: A Python framework for fast computation of mathematical expressions. Preprint at https://arxiv.org/abs/1605.02688 (2016).
Glorot, X. & Bengio, Y. in Proceedings of the Thirteenth International Conference on Artificial Intelligence and Statistics Vol. 9 (eds. Teh, Y.W. & Titterington, M.) 249–256 (Proceedings of Machine Learning Research, 2010).
Kingma, D. & Ba, J. Adam: a method for stochastic optimization. Preprint at https://arxiv.org/abs/1412.6980 (2014).
Schneider, T.D. & Stephens, R.M. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18, 6097–6100 (1990).
Janetzki, S., Cox, J.H., Oden, N. & Ferrari, G. Standardization and validation issues of the ELISPOT assay. Methods Mol. Biol. 302, 51–86 (2005).
Janetzki, S. et al. Guidelines for the automated evaluation of Elispot assays. Nat. Protoc. 10, 1098–1115 (2015).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
DePristo, M.A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
Garrison, E. & Marth, G. Haplotype-based variant detection from short-read sequencing. Preprint at https://arxiv.org/abs/1207.3907 (2012).
Cingolani, P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6, 80–92 (2012).
Szolek, A. et al. OptiType: precision HLA typing from next-generation sequencing data. Bioinformatics 30, 3310–3316 (2014).
Cibulskis, K. et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 31, 213–219 (2013).
Scholz, E.M. et al. Human leukocyte antigen (HLA)-DRB1*15:01 and HLA-DRB5*01:01 present complementary peptide repertoires. Front. Immunol. 8, 984 (2017).
Ooi, J.D. et al. Dominant protection from HLA-linked autoimmunity by antigen-specific regulatory T cells. Nature 545, 243–247 (2017).
Acknowledgements
We would like to thank C.J. Couter for her assistance with general laboratory tasks and establishment of the in vitro stimulation assays. T.A.C. acknowledges funding in part through the NIH/NCI Cancer Center Support Grant P30 CA008748, Pershing Square Sohn Cancer Research grant, the PaineWebber Chair, Stand Up 2 Cancer, NIH R01 CA205426, NIH R35 CA232097, and the STARR Cancer Consortium. V.T.D.M., O.M., G.S., P.B., S.N., N.K., R. Rosell, I.A., N.G., J.H., C.L., K. Choquette, A.S., E.F. and M.F. received research funding support for this study from Gritstone Oncology, Inc.
Author information
Authors and Affiliations
Contributions
Conception and design: B.B.-S., J.B., J.F., M.S., R.Y. Development of methodology: B.B.-S., J.B., J.F., M.S., R.Y., C.D.P., M.J.D., A.C., M.B., L.Y., T.B., V.M., R.Y. Provided patient material and clinical input: V.T.D.M., O.M., G.S., P.B., S.N., N.K., R. Rosell, I.A., N.G., J.H., C.L., A.S., E.F., M.F. Operational support and data management for patient material: K.C., J.A., C.V., K.C. Performed experiments: J.B., C.D.P., M.J.D., T.M., F.D., A.Y., N.C.O., M.G.H., M.S., J.F. Analysis and interpretation of data: B.B.-S., J.B., C.D.P., M.J.D., A.C., M.B., L.Y., T.B., K.J., M.S., J.F., R.Y., N.A.R., T.A.C. Writing, review and/or revision of the manuscript: B.B.-S., J.B., J.F., M.S., R.Y., C.D.P., N.A.R. Study supervision: R.Y., K.J., R. Rousseau.
Corresponding author
Ethics declarations
Competing interests
B.B.-S., J.B., C.D.P., M.J.D., T.M., A.C., M.B., F.D., A.Y., L.Y., N.C.O., K. Caldwell, J.A., T.B., M.G.H., R. Rousseau, C.V., K.J., M.S., J.F. and R.Y. are employees and shareholders of Gritstone Oncology, Inc, a company developing neoantigen immunotherapies. T.A.C. and N.A.R. are founders, shareholders, and serve on the scientific advisory board of Gritstone Oncology. B.B.-S., J.B., C.D.P., T.B., M.S., J.F. and R.Y are inventors on patents and patent applications relating to this work. T.A.C. holds equity in An2H. T.A.C. acknowledges grant funding from Bristol-Myers Squibb, AstraZeneca, Illumina, Pfizer, An2H and Eisai. T.A.C. has served as an advisor for Bristol-Myers Squibb, AstraZeneca, Illumina, Eisai and An2H. T.A.C., N.A.R. and Memorial Sloan Kettering Cancer Center have a patent filing (PCT/US2015/062208) for the use of tumor mutation burden and HLA for prediction of immunotherapy efficacy, which is licensed to Personal Genome Diagnostics. S.N. is on speaker bureaus for Eli Lilly; Bristol-Myers Squibb; Takeda; Merck, Sharp & Dohme; Boehringer Ingelheim; AstraZeneca; and AbbVie.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1, 2 and 5–13, Supplementary Table 1 and Supplementary Notes 1–3 (PDF 2557 kb)
Supplementary Software
EDGE model code (ZIP 4 kb)
Supplementary Figure 3a
Motifs for HLA-A alleles (PDF 726 kb)
Supplementary Figure 3b
Motifs for HLA-B alleles (PDF 570 kb)
Supplementary Figure 3c
Motifs for HLA-C alleles (PDF 647 kb)
Supplementary Figure 4
Precision-recall curves for all test samples (PDF 245 kb)
Supplementary Data 1
Specimen characteristics and MS + NGS metrics (XLSX 18 kb)
Supplementary Data 2
Model predicts HLA peptide stability (CSV 0 kb)
Supplementary Data 3a
T-cell epitope dataset from studies A, B and D (CSV 317 kb)
Supplementary Data 3b
T-cell epitope dataset from study C (CSV 118 kb)
Supplementary Data 3c
Predicted ranks of mutations with pre-existing CD8 response (CSV 1 kb)
Supplementary Data 4
Peptides tested for T-cell recognition in NSCLC patients (CSV 19 kb)
Supplementary Data 5
Demographics of NSCLC patients (XLSX 15 kb)
Supplementary Data 6
Neoantigen and infectious disease epitopes in IVS control (XLSX 18 kb)
Supplementary Data 7
Neoantigen peptides tested in healthy donors (XLSX 17 kb)
Supplementary Data 8
MSD cytokine multiplex and ELISA assays on ELISpot supernatants from NSCLC neoantigen peptides (XLSX 17 kb)
Supplementary Data 9
RNA expression dataset used for model training and testing (CSV 36237 kb)
Rights and permissions
About this article
Cite this article
Bulik-Sullivan, B., Busby, J., Palmer, C. et al. Deep learning using tumor HLA peptide mass spectrometry datasets improves neoantigen identification. Nat Biotechnol 37, 55–63 (2019). https://doi.org/10.1038/nbt.4313
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.4313
This article is cited by
-
COSMIC-based mutation database enhances identification efficiency of HLA-I immunopeptidome
Journal of Translational Medicine (2024)
-
Informing immunotherapy with multi-omics driven machine learning
npj Digital Medicine (2024)
-
New regulatory thinking is needed for AI-based personalised drug and cell therapies in precision oncology
npj Precision Oncology (2024)
-
Artificial intelligence in the treatment of cancer: Changing patterns, constraints, and prospects
Health and Technology (2024)
-
A shared neoantigen vaccine combined with immune checkpoint blockade for advanced metastatic solid tumors: phase 1 trial interim results
Nature Medicine (2024)