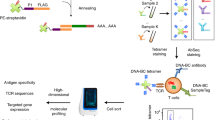
Abstract
We present tetramer-associated T-cell receptor sequencing (TetTCR-seq) to link T cell receptor (TCR) sequences to their cognate antigens in single cells at high throughput. Binding is determined using a library of DNA-barcoded antigen tetramers that is rapidly generated by in vitro transcription and translation. We applied TetTCR-seq to identify patterns in TCR cross-reactivity with cancer neoantigens and to rapidly isolate neoantigen-specific TCRs with no cross-reactivity to the wild-type antigen.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Accession codes
References
Newell, E.W. & Davis, M.M. Nat. Biotechnol. 32, 149–157 (2014).
Bentzen, A.K. et al. Nat. Biotechnol. 34, 1037–1045 (2016).
Strønen, E. et al. Science 352, 1337–1341 (2016).
Glanville, J. et al. Nature 547, 94–98 (2017).
Rodenko, B. et al. Nat. Protoc. 1, 1120–1132 (2006).
Yu, W. et al. Immunity 42, 929–941 (2015).
Zhang, S.Q. et al. Sci. Transl. Med. 8, 341ra377 (2016).
Birnbaum, M.E. et al. Cell 157, 1073–1087 (2014).
Bullock, T.N.J., Mullins, D.W., Colella, T.A. & Engelhard, V.H. J. Immunol. 167, 5824–5831 (2001).
Newell, E.W. et al. Nat. Biotechnol. 31, 623–629 (2013).
Bovay, A. et al. Eur. J. Immunol. 48, 258–272 (2018).
Dietrich, P.-Y. J. Immunol. 170, 5103–5109 (2003).
Cameron, B.J. et al. Sci. Transl. Med. 5, 197ra103 (2013).
Cohen, C.J. et al. J. Clin. Invest. 125, 3981–3991 (2015).
Rajasagi, M. et al. Blood 124, 453–462 (2014).
Carreno, B.M. et al. Science 348, 803–808 (2015).
Nelson, R.W. et al. Immunity 42, 95–107 (2015).
Wilbur, W.J. Mol. Biol. Evol. 2, 434–447 (1985).
Dudley, M.E. et al. Science 298, 850–854 (2002).
Peterson, V.M. et al. Nat. Biotechnol. 35, 936–939 (2017).
Fu, G.K., Wilhelmy, J., Stern, D., Fan, H.C. & Fodor, S.P.A. Anal. Chem. 86, 2867–2870 (2014).
Acknowledgements
We thank B. Wendel for discussions and for producing recombinant HLA-A2; M.M. Davis and H. Huang at Stanford University for discussion of the lentiviral transduction protocol and providing a template TCR construct and HLA-A2 construct; W. Uckert at the Max Delbruck Center for Molecular Medicine for sharing the Jurkat 76 cell line; A. Brock at University of Texas Austin for sharing the HEK 293T cell line; J. Lou at the Institute of Biophysics, Chinese Academy of Sciences, for helping with HCV APL prediction; P. Parker, K. Patel and H. Pan for assistance with initial prototyping; and the NIH tetramer center for additional pMHC tetramer reagents. We also thank anonymous blood donors and staff members at We Are Blood for sample collection. This work was supported by NIH grants R00AG040149 (N.J.), S10OD020072 (N.J.) and R33CA225539 (N.J.), by NSF CAREER Award 1653866 (N.J.), by Welch Foundation grant F1785 (N.J.), by the Robert J. Kleberg, Jr. and Helen C. Kleberg Foundation (N.J.) and by National Natural Science Foundation of China major international (regional) joint research project 81220108006 (W.J.) and NSFC-NHMRC joint research grant 81561128016 (W.J.). N.J. is a Cancer Prevention and Research Institute of Texas (CPRIT) Scholar and a Damon Runyon-Rachleff Innovator. S.-Q.Z. is a recipient of Thrust 2000 – Archie W. Straiton Endowed Graduate Fellowship in Engineering No. 1. A.A.S. is a recipient of the Cockrell School of Engineering fellowship and the Thrust 2000 – Mario E. Ramirez Endowed Graduate Fellowship in Engineering.
Author information
Authors and Affiliations
Contributions
S.-Q.Z. conceived and developed the technology platform. S.-Q.Z and N.J. conceived and designed the study. S.-Q.Z. and K.-Y.M. designed, performed and analyzed data for the majority of experiments; A.A.S. and M.Z. performed TCR cloning, transduction and pMHC tetramer staining studies; C.H. wrote the script for converting sequencing data into TCR sequences, DNA-BC and MIDs, and predicted HCV APLs; C.M.W. and E.S. performed in vitro cell culture and functionality experiments; W.J. cosupervised study and codesigned some experiments; N.J. supervised the study; S.-Q.Z. and N.J. wrote the manuscript with feedback from all authors.
Corresponding authors
Ethics declarations
Competing interests
N.J. is a scientific advisor for ImmuDX LLC and Immune Arch Inc. A provisional patent application has been filed by the University of Texas at Austin for the method described here.
Supplementary information
Supplementary Text and Figures
Supplementary Note and Supplementary Figures 1–20 (PDF 4253 kb)
Supplementary Table 1
Peptide library name, sequence, and fluorescent encoding for each of the six main experiments (XLSX 29 kb)
Supplementary Table 2
TetTCR-seq summary for experiment 1 (XLSX 20 kb)
Supplementary Table 3
TetTCR-Seq summary for experiment 2 (XLSX 35 kb)
Supplementary Table 4
Description of neoantigen and wild-type peptides used for experiments 3 and 4 (XLSX 11 kb)
Supplementary Table 5
TetTCR-seq summary for experiment 3 (XLSX 29 kb)
Supplementary Table 6
TetTCR-seq summary for experiment 4 (XLSX 28 kb)
Supplementary Table 7
Description of neoantigen and wild-type peptides used for experiments 5 and 6 (XLSX 23 kb)
Supplementary Table 8
TetTCR-seq summary for experiment 5 (XLSX 72 kb)
Supplementary Table 9
TetTCR-seq summary for experiment 6 (XLSX 33 kb)
Supplementary Table 10
Primer and DNA template sequences for generating DNA-BC pMHC tetramers and amplification of the DNA-BC (XLSX 26 kb)
Rights and permissions
About this article
Cite this article
Zhang, SQ., Ma, KY., Schonnesen, A. et al. High-throughput determination of the antigen specificities of T cell receptors in single cells. Nat Biotechnol 36, 1156–1159 (2018). https://doi.org/10.1038/nbt.4282
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.4282
This article is cited by
-
Novel insights into TCR-T cell therapy in solid neoplasms: optimizing adoptive immunotherapy
Experimental Hematology & Oncology (2024)
-
The physiological interactome of TCR-like antibody therapeutics in human tissues
Nature Communications (2024)
-
Adaptive immune receptor repertoire analysis
Nature Reviews Methods Primers (2024)
-
Harnessing 3D in vitro systems to model immune responses to solid tumours: a step towards improving and creating personalized immunotherapies
Nature Reviews Immunology (2024)
-
Acoustic tweezers for high-throughput single-cell analysis
Nature Protocols (2023)