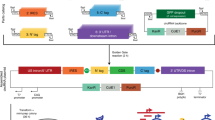
Abstract
Translational regulation by upstream open reading frames (uORFs) is becoming established as a general mechanism for controlling the amount of protein that is synthesized from downstream primary ORFs (pORFs)1,2,3,4,5. We found that genome editing of endogenous uORFs in plants enabled the modulation of translation of mRNAs from four pORFs that are involved in either development or antioxidant biosynthesis. A single-guide RNA that targeted the region harboring a uORF initiation codon can produce multiple mutations. Following uORF editing, we observed varying amounts of mRNA translation in four pORFs. Notably, editing the uORF of LsGGP2, which encodes a key enzyme in vitamin C biosynthesis in lettuce, not only increased oxidation stress tolerance, but also increased ascorbate content by ∼150%. These data indicate that editing plant uORFs provides a generalizable, efficient method for manipulating translation of mRNA that could be applied to dissect biological mechanisms and improve crops.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Calvo, S.E., Pagliarini, D.J. & Mootha, V.K. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc. Natl. Acad. Sci. USA 106, 7507–7512 (2009).
von Arnim, A.G., Jia, Q. & Vaughn, J.N. Regulation of plant translation by upstream open reading frames. Plant Sci. 214, 1–12 (2014).
McGillivray, P. et al. A comprehensive catalog of predicted functional upstream open reading frames in humans. Nucleic Acids Res. 46, 3326–3338 (2018).
Liang, X.H. et al. Translation efficiency of mRNAs is increased by antisense oligonucleotides targeting upstream open reading frames. Nat. Biotechnol. 34, 875–880 (2016).
Xu, G. et al. uORF-mediated translation allows engineered plant disease resistance without fitness costs. Nature 545, 491–494 (2017).
Schwanhäusser, B. et al. Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011).
Pelletier, J., Graff, J., Ruggero, D. & Sonenberg, N. Targeting the eIF4F translation initiation complex: a critical nexus for cancer development. Cancer Res. 75, 250–263 (2015).
Carrieri, C. et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491, 454–457 (2012).
Zucchelli, S., Patrucco, L., Persichetti, F., Gustincich, S. & Cotella, D. Engineering translation in mammalian cell factories to increase protein yield: the unexpected use of long non-coding SINEUP RNAs. Comput. Struct. Biotechnol. J. 14, 404–410 (2016).
Simon, A.E. & Miller, W.A. 3′ cap-independent translation enhancers of plant viruses. Annu. Rev. Microbiol. 67, 21–42 (2013).
Vivinus, S. et al. An element within the 5′ untranslated region of human Hsp70 mRNA which acts as a general enhancer of mRNA translation. Eur. J. Biochem. 268, 1908–1917 (2001).
Pfeiffer, B.D., Truman, J.W. & Rubin, G.M. Using translational enhancers to increase transgene expression in Drosophila. Proc. Natl. Acad. Sci. USA 109, 6626–6631 (2012).
Yin, K., Gao, C. & Qiu, J.L. Progress and prospects in plant genome editing. Nat. Plants 3, 17107 (2017).
Wang, Z.Y., Seto, H., Fujioka, S., Yoshida, S. & Chory, J. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410, 380–383 (2001).
Laing, W.A. et al. An upstream open reading frame is essential for feedback regulation of ascorbate biosynthesis in Arabidopsis. Plant Cell 27, 772–786 (2015).
Hellens, R.P. et al. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1, 13 (2005).
Agius, F. et al. Engineering increased vitamin C levels in plants by overexpression of a D-galacturonic acid reductase. Nat. Biotechnol. 21, 177–181 (2003).
Bulley, S. et al. Enhancing ascorbate in fruits and tubers through over-expression of the L-galactose pathway gene GDP-L-galactose phosphorylase. Plant Biotechnol. J. 10, 390–397 (2012).
Reyes-Chin-Wo, S. et al. Genome assembly with in vitro proximity ligation data and whole-genome triplication in lettuce. Nat. Commun. 8, 14953 (2017).
Qian, W. et al. Molecular and functional analysis of phosphomannomutase (PMM) from higher plants and genetic evidence for the involvement of PMM in ascorbic acid biosynthesis in Arabidopsis and Nicotiana benthamiana. Plant J. 49, 399–413 (2007).
Shan, Q., Wang, Y., Li, J. & Gao, C. Genome editing in rice and wheat using the CRISPR/Cas system. Nat. Protoc. 9, 2395–2410 (2014).
Komor, A.C., Kim, Y.B., Packer, M.S., Zuris, J.A. & Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016).
Gaudelli, N.M. et al. Programmable base editing of A.T to G.C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017).
Hu, J.H. et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57–63 (2018).
Yan, L. et al. High-efficiency genome editing in Arabidopsis using YAO promoter-driven CRISPR/Cas9 system. Mol. Plant 8, 1820–1823 (2015).
Xing, H.L. et al. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14, 327 (2014).
Zhai, Z., Jung, H.I. & Vatamaniuk, O.K. Isolation of protoplasts from tissues of 14-d-old seedlings of Arabidopsis thaliana. J. Vis. Exp. 30, e1149 (2009).
Woo, J.W. et al. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 33, 1162–1164 (2015).
Yoo, S.D., Cho, Y.H. & Sheen, J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572 (2007).
Espinosa-Ruiz, A., Martínez, C. & Prat, S. Protocol to treat seedlings with brassinazole and measure hypocotyl length in Arabidopsis thaliana. Bio Protoc. 5, e1568 (2015).
Cui, F. et al. Arabidopsis ubiquitin conjugase UBC32 is an ERAD component that functions in brassinosteroid-mediated salt stress tolerance. Plant Cell 24, 233–244 (2012).
Zhang, H. et al. The RING finger ubiquitin E3 ligase SDIR1 targets SDIR1-INTERACTING PROTEIN1 for degradation to modulate the salt stress response and ABA signaling in Arabidopsis. Plant Cell 27, 214–227 (2015).
Kovács, L. et al. Quantitative determination of ascorbate from the green alga Chlamydomonas reinhardtii by HPLC. Bio Protoc. 6, e2067 (2016).
Bae, S., Park, J. & Kim, J.S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473–1475 (2014).
Acknowledgements
We thank J.-L. Qui (Institute of Microbiology) for helpful discussion and insightful comments on the manuscript, Y. Wang and G. Wang for advice on the high-performance liquid chromatography assay, and Q. Xie (Institute of Genetics and Developmental Biology) for providing antibodies to BRI1 and PAG. This work was supported by grants from the National Key Research and Development Program of China (2016YFD0101804), the National Natural Science Foundation of China (31788103, 31420103912, 31501376 and 31570369), and the Chinese Academy of Sciences (QYZDY-SSW-SMC030 and GJHZ1602).
Author information
Authors and Affiliations
Contributions
H.Z., X.S., X.J. and C.G. designed the experiments. X.S., H.Z., X.J., R.F. and J.L. performed the experiments. X.J. and K.C. analyzed the results. C.G. supervised the project and C.G., D.W. and H.Z. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have submitted a patent application (application number 201710976945.0) based on the results reported in this paper.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–14 and Supplementary Tables 1–7 (PDF 7506 kb)
Rights and permissions
About this article
Cite this article
Zhang, H., Si, X., Ji, X. et al. Genome editing of upstream open reading frames enables translational control in plants. Nat Biotechnol 36, 894–898 (2018). https://doi.org/10.1038/nbt.4202
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.4202
This article is cited by
-
Application of genome editing techniques to regulate gene expression in crops
BMC Plant Biology (2024)
-
Biofortification’s contribution to mitigating micronutrient deficiencies
Nature Food (2024)
-
Crop bioengineering via gene editing: reshaping the future of agriculture
Plant Cell Reports (2024)
-
Integrating machine learning and genome editing for crop improvement
aBIOTECH (2024)
-
The role and pathway of VQ family in plant growth, immunity, and stress response
Planta (2024)