Abstract
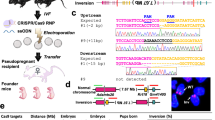
Rapid, efficient generation of knock-in mice with targeted large insertions remains a major hurdle in mouse genetics. Here, we describe two-cell homologous recombination (2C-HR)-CRISPR, a highly efficient gene-editing method based on introducing CRISPR reagents into embryos at the two-cell stage, which takes advantage of the open chromatin structure and the likely increase in homologous-recombination efficiency during the long G2 phase. Combining 2C-HR-CRISPR with a modified biotin–streptavidin approach to localize repair templates to target sites, we achieved a more-than-tenfold increase (up to 95%) in knock-in efficiency over standard methods. We targeted 20 endogenous genes expressed in blastocysts with fluorescent reporters and generated reporter mouse lines. We also generated triple-color blastocysts with all three lineages differentially labeled, as well as embryos carrying the two-component auxin-inducible degradation system for probing protein function. We suggest that 2C-HR-CRISPR is superior to random transgenesis or standard genome-editing protocols, because it ensures highly efficient insertions at endogenous loci and defined 'safe harbor' sites.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Carroll, D. Genome engineering with targetable nucleases. Annu. Rev. Biochem. 83, 409–439 (2014).
Yang, H. et al. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154, 1370–1379 (2013).
Wang, H. et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918 (2013).
Yao, X. et al. Homology-mediated end joining-based targeted integration using CRISPR/Cas9. Cell Res. 27, 801–814 (2017).
Cohen, J. 'Any idiot can do it': genome editor CRISPR could put mutant mice in everyone's reach. Science https://doi.org/10.1126/science.aal0334 (2016).
Chu, V.T. et al. Efficient generation of Rosa26 knock-in mice using CRISPR/Cas9 in C57BL/6 zygotes. BMC Biotechnol. 16, 4 (2016).
Nakade, S. et al. Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat. Commun. 5, 5560 (2014).
Quadros, R.M. et al. Easi-CRISPR: a robust method for one-step generation of mice carrying conditional and insertion alleles using long ssDNA donors and CRISPR ribonucleoproteins. Genome Biol. 18, 92 (2017).
Song, J. et al. RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nat. Commun. 7, 10548 (2016).
Ma, M. et al. Efficient generation of mice carrying homozygous double-floxp alleles using the Cas9-Avidin/Biotin-donor DNA system. Cell Res. 27, 578–581 (2017).
Lee, K. et al. Synthetically modified guide RNA and donor DNA are a versatile platform for CRISPR-Cas9 engineering. eLife 6, e25312 (2017).
Cockburn, K. & Rossant, J. Making the blastocyst: lessons from the mouse. J. Clin. Invest. 120, 995–1003 (2010).
Chen, J. et al. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell 156, 1274–1285 (2014).
Grusch, M. et al. Spatio-temporally precise activation of engineered receptor tyrosine kinases by light. EMBO J. 33, 1713–1726 (2014).
Nishimura, K., Fukagawa, T., Takisawa, H., Kakimoto, T. & Kanemaki, M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods 6, 917–922 (2009).
Hustedt, N. & Durocher, D. The control of DNA repair by the cell cycle. Nat. Cell Biol. 19, 1–9 (2016).
Yang, D. et al. Enrichment of G2/M cell cycle phase in human pluripotent stem cells enhances HDR-mediated gene repair with customizable endonucleases. Sci. Rep. 6, 21264 (2016).
Lin, S., Staahl, B.T., Alla, R.K. & Doudna, J.A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife 3, e04766 (2014).
Gutschner, T., Haemmerle, M., Genovese, G., Draetta, G.F. & Chin, L. Post-translational regulation of Cas9 during G1 enhances homology-directed repair. Cell Reports 14, 1555–1566 (2016).
Natsume, T., Kiyomitsu, T., Saga, Y. & Kanemaki, M.T. Rapid protein depletion in human cells by auxin-inducible degron tagging with short homology donors. Cell Reports 15, 210–218 (2016).
Los, G.V. & Wood, K. The HaloTag: a novel technology for cell imaging and protein analysis. Methods Mol. Biol. 356, 195–208 (2007).
Szymczak, A.L. & Vignali, D.A. Development of 2A peptide-based strategies in the design of multicistronic vectors. Expert Opin. Biol. Ther. 5, 627–638 (2005).
Shcherbakova, D.M. et al. Bright monomeric near-infrared fluorescent proteins as tags and biosensors for multiscale imaging. Nat. Commun. 7, 12405 (2016).
Guo, G. et al. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev. Cell 18, 675–685 (2010).
Posfai, E. et al. Position- and Hippo signaling-dependent plasticity during lineage segregation in the early mouse embryo. eLife 6, e22906 (2017).
Zambrowicz, B.P. et al. Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc. Natl. Acad. Sci. USA 94, 3789–3794 (1997).
Lim, K.H., Huang, H., Pralle, A. & Park, S. Stable, high-affinity streptavidin monomer for protein labeling and monovalent biotin detection. Biotechnol. Bioeng. 110, 57–67 (2013).
Fogarty, N.M.E. et al. Genome editing reveals a role for OCT4 in human embryogenesis. Nature 550, 67–73 (2017).
McDole, K. & Zheng, Y. Generation and live imaging of an endogenous Cdx2 reporter mouse line. Genesis 50, 775–782 (2012).
Strnad, P. et al. Inverted light-sheet microscope for imaging mouse pre-implantation development. Nat. Methods 13, 139–142 (2016).
Royer, L.A. et al. Adaptive light-sheet microscopy for long-term, high-resolution imaging in living organisms. Nat. Biotechnol. 34, 1267–1278 (2016).
Zhang, L., Ward, J.D., Cheng, Z. & Dernburg, A.F. The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development 142, 4374–4384 (2015).
Gu, B., Lambert, J.P., Cockburn, K., Gingras, A.C. & Rossant, J. AIRE is a critical spindle-associated protein in embryonic stem cells. eLife 6, e28131 (2017).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Zernicka-Goetz, M. et al. Following cell fate in the living mouse embryo. Development 124, 1133–1137 (1997).
Grimm, J.B. et al. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods 12, 244–250 (2015).
Cockburn, K., Biechele, S., Garner, J. & Rossant, J. The Hippo pathway member Nf2 is required for inner cell mass specification. Curr. Biol. 23, 1195–1201 (2013).
Acknowledgements
The authors acknowledge technical support from the Model Production Core staff led by M. Gertsenstein at the Centre for Phenogenomics; L. Lavis (HHMI Janelia Research Campus) for synthetic dyes against the Halo-tag; D. Durocher (Lunenfeld-Tanenbaum Research Institute, Mount Sinai Hospital) for discussion of the parameters affecting HR; Y. Yamanaka (McGill Cancer Research Center) for previously setting up the two-cell microinjection system in the Rossant laboratory; and S. Park (University at Buffalo), R. Kuehn (Berlin Institute of Health) and F. Zhang (Broad Institute of Massachusetts Institute of Technology (MIT) and Harvard) for providing materials. This work was funded by CIHR (FDN-143334) to J.R. and Genome Canada and Ontario Genomics (OGI-099).
Author information
Authors and Affiliations
Contributions
B.G., E.P. and J.R. conceived the study. B.G. conceived increasing HR by microinjecting two-cell embryos. B.G. and E.P. designed, carried out and analyzed all experiments equally. J.R. provided supervision and funding for the study. All authors contributed to writing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Validation of correct targeting at targeted locus.
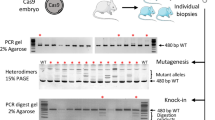
(a) by immunofluorescence staining of 2C-HR-CRISPR microinjected embryos cultured to the blastocyst stage. Embryos were stained for the inserted tag and targeted endogenous gene product (Sox2, Nanog, Oct4, Esrrb, Klf4, Gata6, Gata4, Sox17, Cdx2, Eomes) or where antibodies for the endogenous protein were not available (Fgfr2, Pdgfra, Id2, Dppa1, Gata3) correct lineage localization of the tag was confirmed using established lineage markers. Extended focus images are shown. Scale bars: 40 μm. These experiments were repeated independently 3 times with similar results. (b) by PCR genotyping of Halo-Erk2 and Ctcf-Halo targeting. Single targeted and non-targeted (negative control) embryos were pre-selected by checking for the presence of fluorescently labeled (JF549) Halo-tag at the blastocyst stage and were individually genotyped. These experiments were repeated independently 3 times with similar results.
Supplementary Figure 2 Designs and knock-in efficiencies of targeted alleles.
Table showing all loci targeted, design of targeting construct (C- or N-terminal fusion or C-terminal 2A-fusion), sgRNAs used and targeting efficiencies achieved at the blastocyst stage using 2C-HR-CRISPR (n indicates the number of microinjected embryos, which developed to the blastocyst stage). sgRNA sequence is indicated in the genomic sequence surrounding the insertion site: the N20 bases of the sgRNA are underlined, the PAM sequence is in blue. Start or stop codons in the genomic DNA are highlighted in red or green, respectively. Black arrows show site where the reporter construct was inserted (always after the start codon or before the stop codon). If a synonymous mutation was introduced into the targeting donor to avoid cleavage by the sgRNA/Cas9, this is indicated above the DNA sequence.
Supplementary Figure 3 Analysis of mosaicism in targeted embryos on the basis of immunofluorescence staining data.
(a) Efficiency of correct targeting of knock-in (KI) allele within individual embryos estimated from counting number of reporter positive cells/total number of cells positive for endogenous protein or appropriate lineage marker (where antibody for endogenous protein was not available). Targeting was done using the Cas9/plasmid method. n = number of targeted embryos. Error bars represent s.d. of mean. Results show various levels of KI-mosaicism in individual targeted embryos. (b) Unique mosaicism observed in embryos microinjected with CRISPR reagents to generate Gata6-GFP using the Cas9/plasmid method. We noted that targeting the C-terminus of Gata6 with CRISPR often resulted in a lack of endogenous Gata6 protein in some cells – a unique knock-out phenotype we did not observe with other targeted genes. We suggest this may be due to an indel generated before or at the STOP codon that gives rise to an aberrant transcript or protein that is degraded. Thus in the case of Gata6 targeting we noted mosaicism in microinjected embryos consisting of correct reporter targeting, knock-out and wild-type cells. Examples of different alleles are shown: left embryo likely has wild-type/wild-type (+/+) cells, due to the uniform expression of endogenous Gata6 in PE (stronger) and TE (weaker) cells and no detectable GFP signal. Middle embryo likely has a mix of Gata6-GFP/+ and Gata6-GFP/deleted (-) cells, as GFP is detected in seemingly all Gata6 positive cells, but endogenous Gata6 expression levels vary. Right embryo likely has a mix of +/+, +/- and -/- cells, corresponding to strong, intermediate and completely absent endogenous Gata6 signals. All embryos are from the same microinjection session. Embryos were cultured to the blastocyst stage and immunostained with indicated antibodies. Scale bars: 40 μm. Extended focus images are shown. These experiments were repeated independently 3 times with similar results. (c) Comparison of correct targeting efficiencies of reporter tags in individual embryos using the Cas9/plasmid or Cas9-SA/BIO-PCR methods. Targeting efficiencies were estimated by counting number of reporter positive cells/total number of cells positive for endogenous protein (Nanog-mAID-mCherry and Gata6-Halo) or total cell number marked by DAPI staining (Ctcf-Halo-mAID) (n=number of targeted embryos analyzed). Error bars represent s.d. of mean. p-values were calculated using the Mann-Whitney U-test (2-sided). Results show increased targeting using the Cas9-mSA/BIO-PCR method.
Supplementary Figure 4 Effects of insert size and homology-arm length on targeting efficiency.
Targeting efficiencies were scored in microinjected embryos cultured to the blastocyst stage. n represents number of embryos microinjected. (a) 3 different insert sizes between ~700 to ~1400 bps were tested using both Cas9/plasmid and Cas9-mSA/BIO-PCR methods. Homology arm length was kept constant (3kb:2kb) No obvious decrease in targeting efficiency was observed at this insert size range. (b) 4 different homology arm lengths were compared ranging from 3 kb to 100 bp. Insert size was kept constant (Halo tag, 921 bp). We observed a gradual decrease in targeting efficiencies if the homology arm was shorter than 1 kb.
Supplementary Figure 5 2C-HR-CRISPR targeting efficiencies compared between CD1 and C57BL/6J backgrounds.
Microinjected embryos were cultured to the blastocyst stage where targeting efficiency was evaluated by scoring correct localization of reporter expression (n indicates number of microinjected embryos that developed to the blastocyst stage).
Supplementary Figure 6 Analysis of mosaicism in 2C-HR-CRISPR-targeted blastocysts and mice by genotyping and sequencing.
(a) Schematic overview of strategy to analyze mosaicism in 2C-HR-CRISPR-targeted blastocysts and results of analysis (table). ES cells were derived from Gata6-Halo Cas9/plasmid and Cas9mSA/BIO-PCR microinjected embryos (from 5 knock-in-positive embryos for each injection method). (1) 16 individual ES cell colonies were picked from each derivation and zygosity of the correct knock-in (KI) allele was determined using genotyping PCR. (2) To analyze CRISPR-editing byproduct alleles the targeting region was PCR-amplified, cloned and sequenced (15 bacterial clones from each derivation) from the mixed ES cell population. Wild-type (wt), various deletion (del) and point mutation (point mut) alleles were detected. Last column of table indicates allele types present in each Gata6-Halo-targeted embryo. (b) Schematic overview of strategy to analyze mosaicism in 2C-HR-CRISPR-generated founder mice and results of analysis (table). (3) Presence of KI allele in the germ line of founder animals was assessed by genotyping 2 consecutive N1 litters from each founder (% positive N1 pups (positive pups/total pups)). (4) To analyze CRISPR-editing byproduct alleles the targeting region was PCR-amplified, cloned and sequenced from founder animal tail samples (at least 8 bacterial clones from each founder). Wild-type (wt), various deletion (del) and point mutation (point mut) alleles were detected. Last column of table indicates allele types present in each founder animal.
Supplementary Figure 7 Validations of triple-color reporter blastocysts.
(a) Immunofluorescence staining of N1 blastocysts (all cells are heterozygous, non-mosaic for the reporter) from Nanog-mAID-mCherry and Gata6-Halo mouse lines. These lines were crossed to create the triple-color lineage reporter line. All images show extended focus. Scale bars: 40 μm. These experiments were repeated independently 3 times with similar results. (b) Non-mosaic live triple-color blastocysts with all three lineages labeled with different reporter alleles. Extended focus and single plane image of an early (embryonic day 3.5) and late (embryonic day 4.5) blastocyst are shown. Embryos were produced by breeding 2C-HR-CRISPR-generated mouse lines (Nanog-mAID-mCherry and Gata6-Halo) with an existing Cdx2-eGFP reporter mouse line. The Halo-tag was visualized using the live JF646 dye. Scale bar: 40 μm. These experiments were repeated independently 3 times with similar results.
Supplementary Figure 8 Application of multicolor reporter embryos in live imaging.
Snapshots from time-lapse imaging of non-mosaic Cdx2-eGFP/Gata6-Halo embryos from the 16-cell stage to the blastocyst stage. The Halo-tag was visualized using the live JF549 dye. 1 Z-stack image was acquired every hour for 30 hours with a spinning disk confocal. All images show extended focus. Scale bars: 40 μm. These experiments were repeated independently 3 times with similar results.
Supplementary Figure 9 Additional examples of auxin-inducible degradation of endogenously mAID-tagged protein in early embryos.
Examples of auxin-inducible degradation of Gata6-Halo-mAID and Sox2-mAID-mCherry in preimplantation embryos. Simultaneous targeting of Halo-mAID to the Gata6 locus or mAID-mCherry to the Sox2 locus and TIR1-GFP to the Actb locus was achieved with 2C-HR-CRISPR. Z-stack images were acquired every 10 minutes following addition of IAA. Arrows point to double positive cells with both Gata6-Halo-mAID or Sox2-mAID-mCherry, and TIR1-GFP expression. Gata6-Halo-mAID or Sox2-mAID-mCherry are only degraded in TIR1-GFP positive cells, but not in TIR1-GFP negative cells in the same embryos. Extended focus images are shown. Scale bars: 40 μm. These experiments were repeated independently 3 times with similar results.
Supplementary Figure 10 Schematic overviews of genotyping strategy, donor vectors and BIO-PCR donors.
(a) Schematic overview of strategy to genotype and sequence knock-in reporter alleles, used in mice and ES cells. (b) Schematic overview of strategy to PCR-amplify and sequence editing byproduct alleles, used in mice and ES cells. (c) Schematic overview of EasyFusion targeting vectors. All plasmids are available from Addgene. MCS – multiple cloning site; SA – splice acceptor. (d) Schematic overview of pan-BIO primers used to produce BIO-PCR products.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10 and Supplementary Table 1 (PDF 2662 kb)
Supplementary Table 2
Oligo sequences (XLSX 184 kb)
Live imaging of Gata6-Halo/Cdx2-eGFP mouse embryos from 16-cell stage to blastocyst
Live imaging of mouse embryos carrying primitive endoderm reporter Gata6-Halo and Trophectoderm reporter Cdx2-eGFP for 30 hours 16-cell stage to blastocyst (MOV 5697 kb)
Rights and permissions
About this article
Cite this article
Gu, B., Posfai, E. & Rossant, J. Efficient generation of targeted large insertions by microinjection into two-cell-stage mouse embryos. Nat Biotechnol 36, 632–637 (2018). https://doi.org/10.1038/nbt.4166
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.4166
This article is cited by
-
Donor template delivery by recombinant adeno-associated virus for the production of knock-in mice
BMC Biology (2024)
-
Efficient prime editing in two-cell mouse embryos using PEmbryo
Nature Biotechnology (2024)
-
Various repair events following CRISPR/Cas9-based mutational correction of an infertility-related mutation in mouse embryos
Journal of Assisted Reproduction and Genetics (2024)
-
Genetically modified mice as a tool for the study of human diseases
Molecular Biology Reports (2024)
-
Targeted Gene Insertion: The Cutting Edge of CRISPR Drug Development with Hemophilia as a Highlight
BioDrugs (2024)