Abstract
We report rapid, potent reversal of GalNAc-siRNA-mediated RNA interference (RNAi) activity in vivo with short, synthetic, high-affinity oligonucleotides complementary to the siRNA guide strand. We found that 9-mers with five locked nucleic acids (LNAs) have the highest potency across several targets. Our modular, sequence-specific approach, named REVERSIR, may enhance the therapeutic profile of any long-acting GalNAc–siRNA (short interfering RNA) conjugate by enabling control of RNAi pharmacology.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Accession codes
References
Coelho, T. et al. N. Engl. J. Med. 369, 819–829 (2013).
Fitzgerald, K. et al. Lancet 383, 60–68 (2014).
Fitzgerald, K. et al. N. Engl. J. Med. 376, 41–51 (2017).
Zimmermann, T.S. et al. Mol. Ther. 25, 71–78 (2017).
Nair, J.K. et al. Nucleic Acids Res. 45, 10969–10977 (2017).
Foster, D.J. et al. Mol. Ther. 26, 708–717 (2018).
Meister, G., Landthaler, M., Dorsett, Y. & Tuschl, T. RNA 10, 544–550 (2004).
Hutvágner, G., Simard, M.J., Mello, C.C. & Zamore, P.D. PLoS Biol. 2, e98 (2004).
van der Ree, M.H. et al. Lancet 389, 709–717 (2017).
Rottiers, V. et al. Sci. Transl. Med. 5, 212ra162 (2013).
Janssen, H.L.A. et al. N. Engl. J. Med. 368, 1685–1694 (2013).
Krützfeldt, J. et al. Nature 438, 685–689 (2005).
Crosby, J.R. et al. Nucleic Acid Ther. 25, 297–305 (2015).
Rusconi, C.P. et al. Nat. Biotechnol. 22, 1423–1428 (2004).
Nair, J.K. et al. J. Am. Chem. Soc. 136, 16958–16961 (2014).
Schirle, N.T., Sheu-Gruttadauria, J. & MacRae, I.J. Science 346, 608–613 (2014).
Obad, S. et al. Nat. Genet. 43, 371–378 (2011).
Neben, S. et al. The 51st International Liver Congress, EASL, Barcelona, Spain http://www.natap.org/2016/EASL/EASL_158.htm (2016).
Janas, M.M. et al. Nat. Commun. 9, 723 (2018).
Aronesty, E. https://expressionanalysis.github.io/ea-utils/ (2011).
Dobin, A. et al. Bioinformatics 29, 15–21 (2013).
Liao, Y., Smyth, G.K. & Shi, W. Bioinformatics 30, 923–930 (2014).
Love, M.I., Huber, W. & Anders, S. Genome Biol. 15, 550 (2014).
Elkayam, E. et al. Nucleic Acids Res. 45, 3528–3536 (2017).
Pei, Y. et al. RNA 16, 2553–2563 (2010).
Chen, C. et al. Nucleic Acids Res. 33, e179 (2005).
Rajeev, K.G. et al. ChemBioChem 16, 903–908 (2015).
Edgar, R., Domrachev, M. & Lash, A.E. Nucleic Acids Res. 30, 207–210 (2002).
Acknowledgements
We thank J. Maraganore, R. Meyers, K. Charisse, and K. Fitzgerald for helpful discussions. We thank J. Barry for help with RNA-seq.
Author information
Authors and Affiliations
Contributions
I.Z., A.C., M.M., A.A., K.G.R., M.A.M., and V.J. designed the research. I.Z., S.W., and M.K.S. synthesized oligonucleotides and generated Tm data. A.C. generated in vitro data in PMHs. J.Q. generated in vivo data. R.D. and S.G. generated ASGPR binding and uptake data. C.R.B. quantified siRNA levels and REVERSIR association with Ago2. S.S.-M. generated and H.X. analyzed RNA-seq data. S.M. contributed reagents. I.Z. and V.J. wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
All authors are or were employees of Alnylam Pharmaceuticals when they contributed to this work. REVERSIR is a trademark of Alnylam Pharmaceuticals.
Integrated supplementary information
Supplementary Figure 2 Impact of number and position of Locked Nucleic Acid (LNA) modifications on REVERSIR activity.
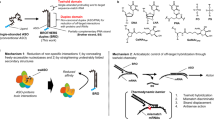
(a) Schematic representation of 15-mer REVERSIR molecules of varying LNA content designed to start with position complementary to the guide strand position g2. Neither the 3′-DNA-A nucleotide linker nor the GalNAc ligand are shown in the schematic or included in the total length assignment of REVERSIR. Complementary nucleotide positions of the guide strand are indicated. All internucleotide linkages are phosphorothioate linkages. Chemistries are represented as follows: OMe: 2′-O-Methyl, PS: phosphorothioate. List of compounds in Supplementary Table 2, R1-R14. (b) In vitro reversal of siRNA mediated knockdown of F7 mRNA by complementary REVERSIR molecules in primary mouse hepatocytes. REVERSIR compounds (100 nM, with titration of 0 – 6 LNA nucleotides) were added to the cells 4 h after transfection of siRNAs (Lipofectamine RNAiMAX transfection, 1 nM). Individual points and mean center plotted. Error bars are s.d. (n=4). (c) Same as (b) but for Ttr target. (d) Positional impact of LNA modification on REVERSIR activity. In vitro reversal of F9-siRNA knockdown activity by 15-mer REVERSIR containing single LNA nucleotide at (compounds R15-R29, Supplementary Table 2) complementary positions opposite to guide nucleotides g2-g16 (the 3′-GalNAc ligand and DNA-A linker were present on all REVERSIR molecules but not shown in the schematic). REVERSIR compounds (100 nM) were added to the primary mouse hepatocytes 4 h after transfection of the F9-siRNA (Lipofectamine RNAiMAX transfection, 1 nM) and after a 48 h incubation, cells were lysed and processed for RNA isolation, cDNA synthesis, and quantitative PCR analysis. F9 mRNA levels were normalized relative to mouse Gapdh mRNA. Individual points and mean center plotted. Error bars are s.d. (n=4).
Supplementary Figure 3 Impact of REVERSIR length on siRNA activity in vivo.
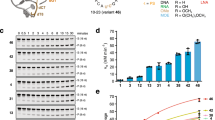
(a) In vivo reversal of F9-siRNA (3 mg/kg) knockdown activity in mice using a 0.75 molar equivalent dose of F9-REVERSIR molecules (22-mer- R-42, 15-mer- R-43, 8-mer- R-45) differing in lengths (normalized to the molar equivalent of the 1.0 mg/kg dose of 15-mer REVERSIR). Measure of center is mean. Error bars are s.d. (n=3). (b) In vivo reversal of TTR-siRNA (3 mg/kg) knockdown activity in mice using a 0.075 molar equivalent dose of TTR-REVERSIR molecules (22-mer- R-6, 15-mer- R-34, 9-mer- R-35) differing in lengths (normalized to the molar equivalent of the 0.1 mg/kg dose of 15-mer REVERSIR). Measure of center is mean. Error bars are s.d. (n=3).
Supplementary Figure 4 Impact of REVERSIR length on in vitro activity by different modes of delivery and Tm.
(a) REVERSIR compounds (22-mer- R-42, 15-mer- R-43, 8-mer- R-45; three-fold serial dilutions from 100 nM – 0.05 nM) were added to the primary mouse hepatocytes 24 h after transfection (Lipofectamine RNAiMAX) of the F9-siRNA (1 nM) and F9 mRNA levels were analyzed by qPCR relative to mouse Gapdh mRNA (48 h after REVERSIR treatment). Measure of center is mean. Error bars are s.d. (n=4). IC50 values are calculated as the concentration corresponding to 50% of target remaining (calculated using XLfit). (b) Same as in (a) but with co-transfection of F9-siRNA and REVERSIRs Lipofectamine RNAiMAX and F9 mRNA analysis 24 h treatment. Measure of center is mean. Error bars are s.d. (n=4). (c) Tm value of each REVERSIR measured after annealing with the complementary F9-siRNA guide strand. Average of two independent annealing sample preparations and measures with difference no more than 0.5 °C (n=2).
Supplementary Figure 5 Impact of REVERSIR length on ASGPR binding, uptake and activity in vitro.
(a) A flow cytometry-based competitive binding assay using a GalNAc-conjugated Alexa647-labeled siRNA substrate was used [Reference 28: Rajeev, K.G. et al. ChemBioChem 16, 903-908 (2015)], where competing unlabeled REVERSIR molecules (22-mer- R-6, 15-mer- R-34, 9-mer- R-35) were incubated at an appropriate range of final concentrations from 3 μM to 0.5 nM in primary mouse hepatocytes (PMH). Fluorescent intensity of the Alexa647 siRNA was quantified to determine the Ki values for ASGPR binding. Individual points and mean center value are plotted. Error bars are s.d. (n=4). The statistical analysis (compared to the data of 9-mer REVERSIR) was performed using multiple t-test one per row (one-sided). (b) Cellular free uptake of Cy5-9-mer (R-66), Cy5-15-mer (R-65) and Cy5-22-mer (R-64) as analyzed by flow cytometry after incubating with PMH for 48 h. Each compound was run in a cell culture plate from three independent wells and each bar represents the mean of samples from three replicate wells. Individual points and mean center value plotted. Error bars are s.d. (n=3). The statistical analysis (compared to the data of 9-mer REVERSIR for each concentration point) was performed using multiple t-test one per row (one-sided). (c) Flow cytometry contour plots analyzed by FloJo software for free uptake of Cy5-labeled REVERSIR molecules of different length utilizing the GalNAc-ASGPR pathway in PMH. The experiment was performed independently twice with similar results. (d) In vitro reversal of TTR-siRNA mRNA knockdown activity using Cy5-labeled REVERSIR molecules of different length. REVERSIR compounds were added for free uptake by PMH for 4 h after transfection (Lipofectamine RNAiMAX) of the TTR-siRNA (1 nM) and Ttr mRNA levels relative to Gapdh mRNA were analyzed 48 h after REVERSIR treatment. Each sample was run in a cell culture plate from three independent wells, and each point represents the mean of three independent wells. Individual points and mean center value are plotted. Error bars are s.d. (n=3). The statistical analysis (compared to the data of 9-mer REVERSIR for each concentration point) was performed using multiple t-test one per row (one-sided).
Supplementary Figure 6 Seed-match coverage and nucleotide tile shift of 15-mer REVERSIR.
REVERSIR compounds (three-fold serial dilutions from 100 nM – 0.05 nM) were added for free uptake by PMH for 4 h after transfection of 1 nM Factor 9-siRNA (Lipofectamine RNAiMAX). F9 mRNA levels relative to Gapdh mRNA were analyzed 48 h after REVERSIR treatment. Each sample was run in a cell culture plate from four independent wells, and each point represents the mean of four independent wells. Mean center value are plotted. Error bars are s.d. (n=4).
Supplementary Figure 7 In vivo reversal of TTR-siRNA activity in mice using 9-mer REVERSIR molecules with different design features.
REVERSIR compounds (0.03 mg/kg) were dosed 7 days after TTR-siRNA (day 0). The serum TTR levels were analyzed at different time points to assess the RNAi activity. R-35: Optimal 9-mer; R-59: Non-cleavable linker; R-60: Non-GalNAc; R-61: Non-targeting. Measure of center is mean. Error bars are s.d. (n=3).
Supplementary Figure 8 Impact of REVERSIR treatment on cellular RNA levels as analyzed by deep sequencing.
Data is shown as volcano plots of RNA sequencing analysis of differentially expressed genes (DEGs). Total RNA isolated from primary mouse hepatocytes 24 h after free uptake incubation with 1 nM TTR-siRNA and 10 nM targeting 9-mer (R-35) or scrambled (SCRA, R-61) TTR-REVERSIR. (a) DEGs analysis of siRNA vs. PBS control: On-target mRNA (Ttr) knockdown is highlighted with a blue asterisk. (b) DEGs analysis of TTR-siRNA + scrambled TTR-REVERSIR vs. PBS control. (c) DEGs analysis of TTR-siRNA + targeting TTR-REVERSIR vs. PBS control. (d, e) DEGs analysis of individual REVERSIR groups vs. PBS control. (f) Ttr mRNA levels by deep sequencing. Individual replicates shown (n=3). Sample size is n = 3. DESeq2 p-values were calculated from a two-sided Wald test on GLM coefficients estimated using a Negative Binomial model [Reference 24: Love, M.I. et al. Genome Biology 15, 550 (2014)], and adjusted for multiple testing using the method of Benjamini and Hochberg.
Supplementary Figure 9 In vivo impact of REVERSIR length on siRNA levels in mouse liver and in immunoprecipitated Argonuate 2 (Ago2).
Animals were SC dosed with targeting REVERSIR or scrambled REVERSIR (SCRA, R-61) at indicated doses 3 days after TTR-siRNA (10 mg/kg) and liver tissue was harvested at day 7 for analysis. (a, d, g) Reversal of TTR-siRNA knockdown activity as measured by Ttr mRNA liver levels normalized to Gapdh mRNA. Individual animal values are shown, center is the mean and error bars are s.d. (n=3). (b, e, h) siRNA levels in total liver lysate were measured by quantifying antisense (guide) strand (μg/g) by RT-qPCR. Individual animal values are shown, center is the mean and error bars are s.d. (n=3). (c, f, i) Levels (ng/g) of TTR-siRNA antisense (guide, black) and sense (passenger, grey) strands in immunoprecipitated Ago2. Individual animal values are shown, center is the mean and error bars are s.d. (n=3). Note that (d, f) are also shown in the main text as Figure 2c, 2d. They are also included here for showing a direct comparison of REVERSIR length impact on Ttr mRNA knockdown and TTR-siRNA loading into Ago2.
Supplementary Figure 10 Uncropped gel images from Northern and Western blots.
(a, b) Uncropped Northern blot images of membranes blotted with an antisense probe (a) or REVERSIR probe (b) matching with Figure 2e of the main text. (c) Western blot analysis of Ago2 IPs. Ago2 band intensities were quantified and normalized to the PBS control group (set to 1). (d) Uncropped Western blot image of Ago2 IPs, matching part (c) of this figure. The Northern and Western experiments were performed independently twice with similar results.
Supplementary Figure 11 Potent and generalizable REVERSIR design evaluated in vivo across three different targets.
(a-c) In vivo reversal of siRNA knockdown activity for three different targets in mice by SC dosed REVERSIR molecules (9-mer with five LNAs) seven days after GalNAc-siRNA treatment. (a) F7 serum activity post F7-siRNA treatment +/− F7-REVERSIR (R-39). (b) TTR serum protein levels post TTR-siRNA treatment +/− TTR-REVERSIR (R-35). (c) F9 serum activity post F9-siRNA treatment +/− F9-REVERSIR (R-44). Dotted grey trace represents no REVERSIR treatment groups. Measure of center is mean. Error bars are s.d. (n=3). (d) In vivo duration of siRNA activity upon REVERSIR treatment. In vivo reversal of 3 mg/kg TTR-siRNA knockdown activity in mice using a 9-mer TTR-REVERSIR (R-35) administered as a single SC dose on day seven at three different doses: 0.1, 0.03 and 0.01 mg/kg. Different doses allow for various degrees of resumption of TTR silencing after a re-challenge with single dose of TTR-siRNA (3 mg/kg) in the same animal groups on day forty-three (thirty-six days post-REVERSIR dose). Measure of center is mean. Error bars are s.d. (n=3).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–11 (PDF 1401 kb)
Supplementary Tables
Supplementary tables1–3 (PDF 229 kb)
Rights and permissions
About this article
Cite this article
Zlatev, I., Castoreno, A., Brown, C. et al. Reversal of siRNA-mediated gene silencing in vivo. Nat Biotechnol 36, 509–511 (2018). https://doi.org/10.1038/nbt.4136
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.4136
This article is cited by
-
RNAi-based drug design: considerations and future directions
Nature Reviews Drug Discovery (2024)
-
RNA interference in the era of nucleic acid therapeutics
Nature Biotechnology (2024)
-
Novel and future lipid-modulating therapies for the prevention of cardiovascular disease
Nature Reviews Cardiology (2023)
-
RNAi-mediated rheostat for dynamic control of AAV-delivered transgenes
Nature Communications (2023)
-
Augmented lipid-nanoparticle-mediated in vivo genome editing in the lungs and spleen by disrupting Cas9 activity in the liver
Nature Biomedical Engineering (2022)