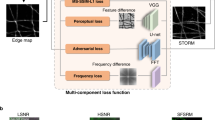
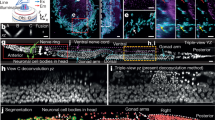
Abstract
The speed of super-resolution microscopy methods based on single-molecule localization, for example, PALM and STORM, is limited by the need to record many thousands of frames with a small number of observed molecules in each. Here, we present ANNA-PALM, a computational strategy that uses artificial neural networks to reconstruct super-resolution views from sparse, rapidly acquired localization images and/or widefield images. Simulations and experimental imaging of microtubules, nuclear pores, and mitochondria show that high-quality, super-resolution images can be reconstructed from up to two orders of magnitude fewer frames than usually needed, without compromising spatial resolution. Super-resolution reconstructions are even possible from widefield images alone, though adding localization data improves image quality. We demonstrate super-resolution imaging of >1,000 fields of view containing >1,000 cells in ∼3 h, yielding an image spanning spatial scales from ∼20 nm to ∼2 mm. The drastic reduction in acquisition time and sample irradiation afforded by ANNA-PALM enables faster and gentler high-throughput and live-cell super-resolution imaging.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Betzig, E. et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 (2006).
Rust, M.J., Bates, M. & Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–796 (2006).
Legant, W.R. et al. High-density three-dimensional localization microscopy across large volumes. Nat. Methods 13, 359–365 (2016).
Deschout, H. et al. Precisely and accurately localizing single emitters in fluorescence microscopy. Nat. Methods 11, 253–266 (2014).
Jones, S.A., Shim, S.H., He, J. & Zhuang, X. Fast, three-dimensional super-resolution imaging of live cells. Nat. Methods 8, 499–505 (2011).
Huang, F. et al. Video-rate nanoscopy using sCMOS camera-specific single-molecule localization algorithms. Nat. Methods 10, 653–658 (2013).
Carlton, P.M. et al. Fast live simultaneous multiwavelength four-dimensional optical microscopy. Proc. Natl. Acad. Sci. USA 107, 16016–16022 (2010).
Stelzer, E.H.K. Light-sheet fluorescence microscopy for quantitative biology. Nat. Methods 12, 23–26 (2015).
Huang, F., Schwartz, S.L., Byars, J.M. & Lidke, K.A. Simultaneous multiple-emitter fitting for single molecule super-resolution imaging. Biomed. Opt. Express 2, 1377–1393 (2011).
Burnette, D.T., Sengupta, P., Dai, Y., Lippincott-Schwartz, J. & Kachar, B. Bleaching/blinking assisted localization microscopy for superresolution imaging using standard fluorescent molecules. Proc. Natl. Acad. Sci. USA 108, 21081–21086 (2011).
Simonson, P.D., Rothenberg, E. & Selvin, P.R. Single-molecule-based super-resolution images in the presence of multiple fluorophores. Nano Lett. 11, 5090–5096 (2011).
Zhu, L., Zhang, W., Elnatan, D. & Huang, B. Faster STORM using compressed sensing. Nat. Methods 9, 721–723 (2012).
Cox, S. et al. Bayesian localization microscopy reveals nanoscale podosome dynamics. Nat. Methods 9, 195–200 (2012).
Ram, S., Ward, E.S. & Ober, R.J. Beyond Rayleigh's criterion: a resolution measure with application to single-molecule microscopy. Proc. Natl. Acad. Sci. USA 103, 4457–4462 (2006).
LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature 521, 436–444 (2015).
Michael, E. Sparse and Redundant Representations: from Theory to Applications in Signal and Image Processing (Springer, 2010).
Hinton, G.E. & Salakhutdinov, R.R. Reducing the dimensionality of data with neural networks. Science 313, 504–507 (2006).
Schmidhuber, J. Deep learning in neural networks: an overview. Neural Netw. 61, 85–117 (2015).
Sage, D. et al. Quantitative evaluation of software packages for single-molecule localization microscopy. Nat. Methods 12, 717–724 (2015).
Isola, P., Zhu, J.-Y., Zhou, T. & Efros, A.A. Image-to-image translation with conditional adversarial networks. Preprint at http://arxiv.org/abs/1611.07004 (2016).
Ronneberger, O., Fischer, P. & Brox, T. in Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. (eds., Navab, N., Hornegger, J., Wells, W.M. & Frangi, A.F.), 234–241 (Springer; 2015).
Goodfellow, I. et al. Generative adversarial nets. Adv. Neural Inf. Process. Syst. 27, 2672–2680 (2014).
Ciresan, D.C., Giusti, A., Gambardella, L.M. & Schmidhuber, J. Mitosis detection in breast cancer histology images with deep neural networks. Med. Image Comput. Comput. Assist. Interv. 2013, 8150 (2013).
Wang, Z.W.Z. & Bovik, A.C.A.C. Mean squared error: love it or leave it? A new look at Signal Fidelity Measures. IEEE Signal Process. Mag. 26, 98–117 (2009).
Zhao, H., Gallo, O., Frosio, I. & Kautz, J. Loss functions for image restoration with neural networks. IEEE Trans. Comput. IMAGING 3, 47–57 (2017).
Culley, S. et al. Quantitative mapping and minimization of super-resolution optical imaging artifacts. Nat. Methods 15, 263–266 (2018).
Srivastava, N., Hinton, G., Krizhevsky, A., Sutskever, I. & Salakhutdinov, R. Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 15, 1929–1958 (2014).
Arbona, J.-M., Herbert, S., Fabre, E. & Zimmer, C. Inferring the physical properties of yeast chromatin through Bayesian analysis of whole nucleus simulations. Genome Biol. 18, 81 (2017).
Arnal, I. & Wade, R.H. How does taxol stabilize microtubules? Curr. Biol. 5, 900–908 (1995).
Wu, S. et al. Microtubule motors regulate ISOC activation necessary to increase endothelial cell permeability. J. Biol. Chem. 282, 34801–34808 (2007).
De Brabander, M., De May, J., Joniau, M. & Geuens, G. Ultrastructural immunocytochemical distribution of tubulin in cultured cells treated with microtubule inhibitors. Cell Biol. Int. Rep. 1, 177–183 (1977).
Schnitzbauer, J., Strauss, M.T., Schlichthaerle, T., Schueder, F. & Jungmann, R. Super-resolution microscopy with DNA-PAINT. Nat. Protoc. 12, 1198–1228 (2017).
Löschberger, A. et al. Super-resolution imaging visualizes the eightfold symmetry of gp210 proteins around the nuclear pore complex and resolves the central channel with nanometer resolution. J. Cell Sci. 125, 570–575 (2012).
Szymborska, A. et al. Nuclear pore scaffold structure analyzed by super-resolution microscopy and particle averaging. Science 341, 655–658 (2013).
Sellés, J. et al. Nuclear pore complex plasticity during developmental process as revealed by super-resolution microscopy. Sci. Rep. 7, 14732 (2017).
Bellot, G. et al. TOM22, a core component of the mitochondria outer membrane protein translocation pore, is a mitochondrial receptor for the proapoptotic protein Bax. Cell Death Differ. 14, 785–794 (2007).
Boettiger, A.N. et al. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature 529, 418–422 (2016).
Zhang, Z., Nishimura, Y. & Kanchanawong, P. Extracting microtubule networks from superresolution single-molecule localization microscopy data. Mol. Biol. Cell 28, 333–345 (2017).
Neumann, B. et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature 464, 721–727 (2010).
Beghin, A. et al. Localization-based super-resolution imaging meets high-content screening. Nat. Methods 14, 1184–1190 (2017).
Ouyang, W. & Zimmer, C. The imaging tsunami: computational opportunities and challenges. Curr. Opin. Syst. Biol. 4, 105–113 (2017).
Chen, F., Tillberg, P.W. & Boyden, E.S. Optical imaging. Expansion microscopy. Science 347, 543–548 (2015).
Chang, J.-B. et al. Iterative expansion microscopy. Nat. Methods 14, 593–599 (2017).
de Boer, P., Hoogenboom, J.P. & Giepmans, B.N.G. Correlated light and electron microscopy: ultrastructure lights up!. Nat. Methods 12, 503–513 (2015).
Huang, B., Wang, W., Bates, M. & Zhuang, X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 319, 810–813 (2008).
Bates, M., Huang, B., Dempsey, G.T. & Zhuang, X. Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science 317, 1749–1753 (2007).
Jungmann, R. et al. Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT. Nat. Methods 11, 313–318 (2014).
Shroff, H., Galbraith, C.G., Galbraith, J.A. & Betzig, E. Live-cell photoactivated localization microscopy of nanoscale adhesion dynamics. Nat. Methods 5, 417–423 (2008).
Kendall, A. & Gal, Y. What uncertainties do we need in Bayesian deep learning for computer vision? Adv. Neural Inf. Process. Syst. 30, 5580–5590 (2017).
Xu, L., Choy, C.S. & Li, Y.W. in 2016 IEEE International Workshop on Acoustic Signal Enhancement, IWAENC 2016 (IEEE, 2016) doi:10.1109/IWAENC.2016.7602891.
Maas, A.L., Hannun, A.Y. & Ng, A.Y. Rectifier nonlinearities improve neural network acoustic models. Proc. 30th Int. Conf. Mach. Learn. 30, 3 (2013).
Mao, X. et al. Least squares generative adversarial networks. ICCV 2794–2802 (2017) doi:10.1109/ICCV.2017.304.
Kingma, D.P. & Ba, J. Adam: a method for stochastic optimization. ICLR 1–15. http://doi.acm.org.ezproxy.lib.ucf.edu/10.1145/1830483.1830503 (2015).
Rampasek, L. & Goldenberg, A. TensorFlow: biology's gateway to deep learning? Cell Syst. 2, 12–14 (2016).
Reddy, B.S. & Chatterji, B.N. An FFT-based technique for translation, rotation, and scale-invariant image registration. IEEE Trans. Image Process. 5, 1266–1271 (1996).
Simard, P.Y., Steinkraus, D. & Platt, J.C. Best practices for convolutional neural networks applied to visual document analysis. Seventh Int. Conf. Doc. Anal. Recognition, 2003. Proceedings 1, 958–963 (2003).
Plimpton, S. Fast parallel algorithms for short-range molecular dynamics. J. Comput. Phys. 117, 1–19 (1995).
van de Linde, S. et al. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nat. Protoc. 6, 991–1009 (2011).
Henriques, R. et al. QuickPALM: 3D real-time photoactivation nanoscopy image processing in ImageJ. Nat. Methods 7, 339–340 (2010).
Lelek, M. et al. Superresolution imaging of HIV in infected cells with FlAsH-PALM. Proc. Natl. Acad. Sci. USA 109, 8564–8569 (2012).
Edelstein, A., Amodaj, N., Hoover, K., Vale, R. & Stuurman, N. Computer control of microscopes using μmanager. Curr. Protoc. Mol. Biol. 92, 14.20.1–14.20.17 (2010).
Ovesný, M., Křížek, P., Borkovec, J., Svindrych, Z. & Hagen, G.M. ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics 30, 2389–2390 (2014).
Preibisch, S., Saalfeld, S. & Tomancak, P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics 25, 1463–1465 (2009).
Sergé, A., Bertaux, N., Rigneault, H. & Marguet, D. Dynamic multiple-target tracing to probe spatiotemporal cartography of cell membranes. Nat. Methods 5, 687–694 (2008).
Acknowledgements
We thank the following colleagues for useful discussions and suggestions and/or critical reading of the manuscript: C. Leduc, S. Etienne-Manneville, S. Lévêque-Fort, N. Bourg, A. Echard, J.-B. Masson, T. Rose, P. Hersen, F. Mueller, M. Cohen, Z. Zhang, and P. Kanchanawong. We also thank the four anonymous reviewers for their constructive criticism, which led to significant improvements of ANNA-PALM. We further thank O. Faklaris, J. Sellés and M. Penrad (Institut Jacques Monod), and F. Montel (Ecole Normale Supérieure de Lyon) for providing Xenopus nuclear pore data, J. Bai (Institut Pasteur) for TOM22 antibodies, and C. Leterrier for fixation protocols. We thank E. Rensen and C. Weber for help with experiments and suggestions, B. Lelandais for help with PALM image processing, J.-B. Arbona for polymer simulations and J. Parmar for suggestions that led to the name ANNA-PALM. We thank the IT service of Institut Pasteur, including J.-B. Denis, N. Joly, and S. Fournier, for access to the HPC cluster and relevant assistance, and T. Huynh for help with GPU computing. This work was funded by Institut Pasteur, Agence Nationale de la Recherche grant (ANR 14 CE10 0018 02), Fondation pour la Recherche Médicale (Equipe FRM, DEQ 20150331762), and the Région Ile de France (DIM Malinf). We also acknowledge Investissement d'Avenir grant ANR-16-CONV-0005 for funding a GPU farm used in this work. A.A. and X.H. are recipients of Pasteur-Roux fellowships from Institut Pasteur. W.O. is a scholar in the Pasteur–Paris University (PPU) International PhD program.
Author information
Authors and Affiliations
Contributions
W.O. conceived the method, developed ANNA-PALM software and web application, and performed experiments and analyses. A.A., M.L., and X.H. performed experiments. C.Z. conceived the method, supervised the project, and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
W.O. and C.Z. are listed as inventors on European patent applications EP17306022 and EP18305225.7 filed by Institut Pasteur.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–15, Supplementary Tables 1–4, Supplementary Note 1 Neural network architecture—A-net (PDF 37749 kb)
ANNA-PALM reconstruction quality improves with increasing acquisition time
This video shows an animated version of Figure 3, where the number of frames in the sparse PALM image (b) increases from k=0 to k=30,000. The quality of the corresponding ANNA-PALM reconstruction (e) increases with frame number, as shown by the merged image (h), where the ANNA-PALM reconstruction is shown in red and the dense PALM image obtained from K=30,000 frames (c) is shown in green. Panels a, c, d, f, g, i are identical to the corresponding panels in Figure 3. (MOV 17029 kb)
High-throughput super-resolution imaging with ANNA-PALM
This video shows a zoom-in into a mosaic image covering a 1.8 mm × 1.8 mm field, assembled from 1,089 individual fields of view. Top left: widefield mosaic image assembled from 1,089 widefield images. Top right: sparse PALM mosaic image assembled from 1,089 individual sparse PALM images, each of which was obtained from k=1,000 diffraction limited frames. Bottom left: mosaic of ANNA-PALM images reconstructed from the widefield images alone. Bottom right: mosaic ofANNA-PALM images reconstructed from the widefield images and the sparse PALM images combined. (MP4 26187 kb)
Rights and permissions
About this article
Cite this article
Ouyang, W., Aristov, A., Lelek, M. et al. Deep learning massively accelerates super-resolution localization microscopy. Nat Biotechnol 36, 460–468 (2018). https://doi.org/10.1038/nbt.4106
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.4106
This article is cited by
-
Computational drug development for membrane protein targets
Nature Biotechnology (2024)
-
Content-aware frame interpolation (CAFI): deep learning-based temporal super-resolution for fast bioimaging
Nature Methods (2024)
-
Live-cell imaging powered by computation
Nature Reviews Molecular Cell Biology (2024)
-
SpiDe-Sr: blind super-resolution network for precise cell segmentation and clustering in spatial proteomics imaging
Nature Communications (2024)
-
Enhancing image resolution of confocal fluorescence microscopy with deep learning
PhotoniX (2023)