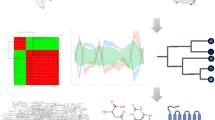
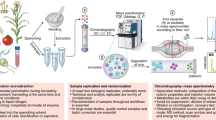
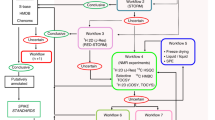
Abstract
Metabolomics, in which small-molecule metabolites (the metabolome) are identified and quantified, is broadly acknowledged to be the omics discipline that is closest to the phenotype1,2,3. Although appreciated for its role in biomarker discovery programs, metabolomics can also be used to identify metabolites that could alter a cell's or an organism's phenotype. Metabolomics activity screening (MAS) as described here integrates metabolomics data with metabolic pathways and systems biology information, including proteomics and transcriptomics data, to produce a set of endogenous metabolites that can be tested for functionality in altering phenotypes. A growing literature reports the use of metabolites to modulate diverse processes, such as stem cell differentiation, oligodendrocyte maturation, insulin signaling, T-cell survival and macrophage immune responses. This opens up the possibility of identifying and applying metabolites to affect phenotypes. Unlike genes or proteins, metabolites are often readily available, which means that MAS is broadly amenable to high-throughput screening of virtually any biological system.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Johnson, C.H., Ivanisevic, J. & Siuzdak, G. Metabolomics: beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 17, 451–459 (2016).
Patti, G.J., Yanes, O. & Siuzdak, G. Metabolomics: the apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 13, 263–269 (2012).
Fiehn, O. Metabolomics--the link between genotypes and phenotypes. Plant Mol. Biol. 48, 155–171 (2002).
Woolf, L.I., Griffiths, R. & Moncrieff, A. Treatment of phenylketonuria with a diet low in phenylalanine. BMJ 1, 57–64 (1955).
Kamanna, V.S. & Kashyap, M.L. Mechanism of action of niacin. Am. J. Cardiol. 101 8A, 20B–26B (2008).
Banach, M. et al. Statin therapy and plasma coenzyme Q10 concentrations--A systematic review and meta-analysis of placebo-controlled trials. Pharmacol. Res. 99, 329–336 (2015).
Tautenhahn, R., Patti, G.J., Rinehart, D. & Siuzdak, G. XCMS Online: a web-based platform to process untargeted metabolomic data. Anal. Chem. 84, 5035–5039 (2012).
Pluskal, T., Castillo, S., Villar-Briones, A. & Orešicčˇ, M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics 11, 395 (2010).
Xia, J., Sinelnikov, I.V., Han, B. & Wishart, D.S. MetaboAnalyst 3.0-making metabolomics more meaningful. Nucleic Acids Res. 43, W251–W257 (2015).
Tautenhahn, R. et al. An accelerated workflow for untargeted metabolomics using the METLIN database. Nat. Biotechnol. 30, 826–828 (2012).
Wishart, D.S. et al. HMDB: the Human Metabolome Database. Nucleic Acids Res. 35, D521–D526 (2007).
Fahy, E. et al. A comprehensive classification system for lipids. J. Lipid Res. 46, 839–861 (2005).
Vinaixa, M. et al. Mass spectral databases for LC/MS- and GC/MS-based metabolomics: State of the field and future prospects. Trends Analyt. Chem. 78, 23–35 (2016).
Yanes, O. et al. Metabolic oxidation regulates embryonic stem cell differentiation. Nat. Chem. Biol. 6, 411–417 (2010).
Sperber, H. et al. The metabolome regulates the epigenetic landscape during naive-to-primed human embryonic stem cell transition. Nat. Cell Biol. 17, 1523–1535 (2015).
Gil-de-Gómez, L. et al. A phosphatidylinositol species acutely generated by activated macrophages regulates innate immune responses. J. Immunol. 190, 5169–5177 (2013).
Guijas, C., Meana, C., Astudillo, A.M., Balboa, M.A. & Balsinde, J. Foamy monocytes are enriched in cis-7-hexadecenoic fatty acid (16:1n-9), a possible biomarker for early detection of cardiovascular disease. Cell Chem. Biol. 23, 689–699 (2016).
Hinz, C. et al. Human platelets utilize cycloxygenase-1 to generate dioxolane A3, a neutrophil-activating eicosanoid. J. Biol. Chem. 291, 13448–13464 (2016).
Geiger, R. et al. L-arginine modulates t cell metabolism and enhances survival and anti-tumor activity. Cell 167, 829–842.e13 (2016).
Beyer, B.A. et al. Metabolomics-based discovery of a metabolite that enhances oligodendrocyte maturation. Nat. Chem. Biol. 14, 22–28 (2018).
Patti, G.J. et al. Metabolomics implicates altered sphingolipids in chronic pain of neuropathic origin. Nat. Chem. Biol. 8, 232–234 (2012).
Yore, M.M. et al. Discovery of a class of endogenous mammalian lipids with anti-diabetic and anti-inflammatory effects. Cell 159, 318–332 (2014).
Prentice, K.J. et al. The furan fatty acid metabolite CMPF is elevated in diabetes and induces β cell dysfunction. Cell Metab. 19, 653–666 (2014).
Huan, T. et al. Systems biology guided by XCMS Online metabolomics. Nat. Methods 14, 461–462 (2017).
Wohlgemuth, G. et al. SPLASH, a hashed identifier for mass spectra. Nat. Biotechnol. 34, 1099–1101 (2016).
Koeth, R.A. et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 19, 576–585 (2013).
Wang, Z. et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57–63 (2011).
Wang, Z. et al. Non-lethal inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis. Cell 163, 1585–1595 (2015).
Wikoff, W.R. et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. USA 106, 3698–3703 (2009).
Panopoulos, A.D. et al. The metabolome of induced pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell Res. 22, 168–177 (2012).
Ezashi, T., Das, P. & Roberts, R.M. Low O2 tensions and the prevention of differentiation of hES cells. Proc. Natl. Acad. Sci. USA 102, 4783–4788 (2005).
Fuhrer, T., Heer, D., Begemann, B. & Zamboni, N. High-throughput, accurate mass metabolome profiling of cellular extracts by flow injection-time-of-flight mass spectrometry. Anal. Chem. 83, 7074–7080 (2011).
Franklin, R.J.M. & Ffrench-Constant, C. Remyelination in the CNS: from biology to therapy. Nat. Rev. Neurosci. 9, 839–855 (2008).
Deshmukh, V.A. et al. A regenerative approach to the treatment of multiple sclerosis. Nature 502, 327–332 (2013).
Fernández-Pisonero, I. et al. Synergy between sphingosine 1-phosphate and lipopolysaccharide signaling promotes an inflammatory, angiogenic and osteogenic response in human aortic valve interstitial cells. PLoS One 9, e109081 (2014).
Cardoso, C., Afonso, C. & Bandarra, N.M. Dietary DHA and health: cognitive function ageing. Nutr. Res. Rev. 29, 281–294 (2016).
Ng, C.M., Blackman, M.R., Wang, C. & Swerdloff, R.S. The role of carnitine in the male reproductive system. Ann. NY Acad. Sci. 1033, 177–188 (2004).
Wise, L.E., Shelton, C.C., Cravatt, B.F., Martin, B.R. & Lichtman, A.H. Assessment of anandamide's pharmacological effects in mice deficient of both fatty acid amide hydrolase and cannabinoid CB1 receptors. Eur. J. Pharmacol. 557, 44–48 (2007).
Hardeland, R. et al. Melatonin—a pleiotropic, orchestrating regulator molecule. Prog. Neurobiol. 93, 350–384 (2011).
Cohen, L.J. et al. Commensal bacteria make GPCR ligands that mimic human signalling molecules. Nature 549, 48–53 (2017).
Cravatt, B.F. et al. Chemical characterization of a family of brain lipids that induce sleep. Science 268, 1506–1509 (1995).
Kosina, S.M. et al. Exometabolomics assisted design and validation of synthetic obligate mutualism. ACS Synth. Biol. 5, 569–576 (2016).
Acknowledgements
We gratefully acknowledge financial support from the National Institutes of Health (Grants R01 GM114368-03, P30 MH062261-10, P01 DA026146-02), and support was also received from Ecosystems and Networks Integrated with Genes and Molecular Assemblies (http://enigma.lbl.gov), a Scientific Focus Area Program at Lawrence Berkeley Laboratory for the US Department of Energy, Office of Science, Office of Biological and Environmental Research under Contract DE-AC02-05CH11231.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Guijas, C., Montenegro-Burke, J., Warth, B. et al. Metabolomics activity screening for identifying metabolites that modulate phenotype. Nat Biotechnol 36, 316–320 (2018). https://doi.org/10.1038/nbt.4101
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.4101