Abstract
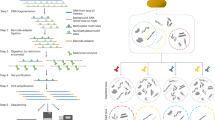
Shotgun metagenomics methods enable characterization of microbial communities in human microbiome and environmental samples. Assembly of metagenome sequences does not output whole genomes, so computational binning methods have been developed to cluster sequences into genome 'bins'. These methods exploit sequence composition, species abundance, or chromosome organization but cannot fully distinguish closely related species and strains. We present a binning method that incorporates bacterial DNA methylation signatures, which are detected using single-molecule real-time sequencing. Our method takes advantage of these endogenous epigenetic barcodes to resolve individual reads and assembled contigs into species- and strain-level bins. We validate our method using synthetic and real microbiome sequences. In addition to genome binning, we show that our method links plasmids and other mobile genetic elements to their host species in a real microbiome sample. Incorporation of DNA methylation information into shotgun metagenomics analyses will complement existing methods to enable more accurate sequence binning.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
BioProject
NCBI Reference Sequence
Referenced accessions
GenBank/EMBL/DDBJ
NCBI Reference Sequence
Sequence Read Archive
References
Cho, I. & Blaser, M.J. The human microbiome: at the interface of health and disease. Nat. Rev. Genet. 13, 260–270 (2012).
Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Janda, J.M. & Abbott, S.L. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J. Clin. Microbiol. 45, 2761–2764 (2007).
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010).
Tyson, G.W. et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428, 37–43 (2004).
Modi, S.R., Lee, H.H., Spina, C.S. & Collins, J.J. Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature 499, 219–222 (2013).
Luo, C. et al. ConStrains identifies microbial strains in metagenomic datasets. Nat. Biotechnol. 33, 1045–1052 (2015).
Kuleshov, V. et al. Synthetic long-read sequencing reveals intraspecies diversity in the human microbiome. Nat. Biotechnol. 34, 64–69 (2016).
Brady, A. & Salzberg, S.L. Phymm and PhymmBL: metagenomic phylogenetic classification with interpolated Markov models. Nat. Methods 6, 673–676 (2009).
Wood, D.E. & Salzberg, S.L. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 15, R46 (2014).
Saeed, I., Tang, S.L. & Halgamuge, S.K. Unsupervised discovery of microbial population structure within metagenomes using nucleotide base composition. Nucleic Acids Res. 40, e34 (2012).
Iverson, V. et al. Untangling genomes from metagenomes: revealing an uncultured class of marine Euryarchaeota. Science 335, 587–590 (2012).
Laczny, C.C., Pinel, N., Vlassis, N. & Wilmes, P. Alignment-free visualization of metagenomic data by nonlinear dimension reduction. Sci. Rep. 4, 4516 (2014).
Laczny, C.C. et al. VizBin - an application for reference-independent visualization and human-augmented binning of metagenomic data. Microbiome 3, 1–7 (2015).
Sharon, I. et al. Time series community genomics analysis reveals rapid shifts in bacterial species, strains, and phage during infant gut colonization. Genome Res. 23, 111–120 (2013).
Albertsen, M. et al. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat. Biotechnol. 31, 533–538 (2013).
Nielsen, H.B. et al. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat. Biotechnol. 32, 822–828 (2014).
Alneberg, J. et al. Binning metagenomic contigs by coverage and composition. Nat. Methods 11, 1144–1146 (2014).
Marbouty, M. et al. Metagenomic chromosome conformation capture (meta3C) unveils the diversity of chromosome organization in microorganisms. eLife 3, e03318 (2014).
Burton, J.N., Liachko, I., Dunham, M.J. & Shendure, J. Species-level deconvolution of metagenome assemblies with Hi-C-based contact probability maps. G3 (Bethesda) 4, 1339–1346 (2014).
Marbouty, M., Baudry, L., Cournac, A. & Koszul, R. Scaffolding bacterial genomes and probing host-virus interactions in gut microbiome by proximity ligation (chromosome capture) assay. Sci. Adv. 3, e1602105 (2017).
Flusberg, B.A. et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat. Methods 7, 461–465 (2010).
Eid, J. et al. Real-time DNA sequencing from single polymerase molecules. Science 323, 133–138 (2009).
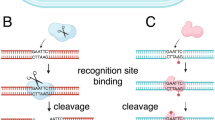
Casadesús, J. & Low, D. Epigenetic gene regulation in the bacterial world. Microbiol. Mol. Biol. Rev. 70, 830–856 (2006).
Blow, M.J. et al. The epigenomic landscape of prokaryotes. PLoS Genet. 12, e1005854 (2016).
Kobayashi, I., Nobusato, A., Kobayashi-Takahashi, N. & Uchiyama, I. Shaping the genome--restriction-modification systems as mobile genetic elements. Curr. Opin. Genet. Dev. 9, 649–656 (1999).
Conlan, S. et al. Single-molecule sequencing to track plasmid diversity of hospital-associated carbapenemase-producing Enterobacteriaceae. Sci. Transl. Med. 6, 254ra126 (2014).
Schadt, E.E. et al. Modeling kinetic rate variation in third generation DNA sequencing data to detect putative modifications to DNA bases. Genome Res. 23, 129–141 (2013).
Beaulaurier, J. et al. Single molecule-level detection and long read-based phasing of epigenetic variations in bacterial methylomes. Nat. Commun. 6, 7438 (2015).
van der Maaten, L. & Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
van der Maaten, L. Accelerating t-sne using tree-based algorithms. J. Mach. Learn. Res. 15, 3221–3245 (2014).
Kim, M., Oh, H.S., Park, S.C. & Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 64, 346–351 (2014).
Parks, D.H., Imelfort, M., Skennerton, C.T., Hugenholtz, P. & Tyson, G.W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055 (2015).
Uchimura, Y. et al. Complete genome sequences of 12 species of Stable Defined Moderately Diverse Mouse Microbiota 2. Genome Announc. 4, e00951–16 (2016).
Ormerod, K.L. et al. Genomic characterization of the uncultured Bacteroidales family S24-7 inhabiting the guts of homeothermic animals. Microbiome 4, 36 (2016).
Xiao, L. et al. A catalog of the mouse gut metagenome. Nat. Biotechnol. 33, 1103–1108 (2015).
Wannemuehler, M.J., Overstreet, A.M., Ward, D.V. & Phillips, G.J. Draft genome sequences of the altered schaedler flora, a defined bacterial community from gnotobiotic mice. Genome Announc. 2, e00287–14 (2014).
Imelfort, M. et al. GroopM: an automated tool for the recovery of population genomes from related metagenomes. PeerJ 2, e603 (2014).
Kang, D.D., Froula, J., Egan, R. & Wang, Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 3, e1165 (2015).
Slater, F.R., Bailey, M.J., Tett, A.J. & Turner, S.L. Progress towards understanding the fate of plasmids in bacterial communities. FEMS Microbiol. Ecol. 66, 3–13 (2008).
Thomas, C.M. & Nielsen, K.M. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3, 711–721 (2005).
Fang, G. et al. Genome-wide mapping of methylated adenine residues in pathogenic Escherichia coli using single-molecule real-time sequencing. Nat. Biotechnol. 30, 1232–1239 (2012).
Roberts, R.J., Vincze, T., Posfai, J. & Macelis, D. REBASE—a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 43, D298–D299 (2015).
Coyne, M.J., Zitomersky, N.L., McGuire, A.M., Earl, A.M. & Comstock, L.E. Evidence of extensive DNA transfer between bacteroidales species within the human gut. MBio 5, e01305–e01314 (2014).
Chin, C.-S. et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569 (2013).
Krebes, J. et al. The complex methylome of the human gastric pathogen Helicobacter pylori. Nucleic Acids Res. 42, 2415–2432 (2014).
Clarke, J. et al. Continuous base identification for single-molecule nanopore DNA sequencing. Nat. Nanotechnol. 4, 265–270 (2009).
Fuller, C.W. et al. Real-time single-molecule electronic DNA sequencing by synthesis using polymer-tagged nucleotides on a nanopore array. Proc. Natl. Acad. Sci. USA 113, 5233–5238 (2016).
Rand, A.C. et al. Mapping DNA methylation with high-throughput nanopore sequencing. Nat. Methods 14, 411–413 (2017).
Lan, F., Demaree, B., Ahmed, N. & Abate, A.R. Single-cell genome sequencing at ultra-high-throughput with microfluidic droplet barcoding. Nat. Biotechnol. 35, 640–646 (2017).
Rousseeuw, P.J. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 20, 53–65 (1987).
Caporaso, J.G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010).
Sokol, H. et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA 105, 16731–16736 (2008).
Livanos, A.E. et al. Antibiotic-mediated gut microbiome perturbation accelerates development of type 1 diabetes in mice. Nat. Microbiol. 1, 16140 (2016).
Heuermann, D. & Haas, R. A stable shuttle vector system for efficient genetic complementation of Helicobacter pylori strains by transformation and conjugation. Mol. Gen. Genet. 257, 519–528 (1998).
Zhang, X.S. & Blaser, M.J. Natural transformation of an engineered Helicobacter pylori strain deficient in type II restriction endonucleases. J. Bacteriol. 194, 3407–3416 (2012).
Leonard, M.T. et al. The methylome of the gut microbiome: disparate Dam methylation patterns in intestinal Bacteroides dorei . Front. Microbiol. 5, 361 (2014).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Feng, Z. et al. Detecting DNA modifications from SMRT sequencing data by modeling sequence context dependence of polymerase kinetic. PLOS Comput. Biol. 9, e1002935 (2013).
Kurtz, S. et al. Versatile and open software for comparing large genomes. Genome Biol. 5, R12 (2004).
Becker, L. et al. Complete genome sequence of a CTX-M-15-producing Klebsiella pneumoniae outbreak strain from multilocus sequence type 514. Genome Announc. 3, e00742–e15 (2015).
Müllner, D. fastcluster: Fast hierarchical, agglomerative. J. Stat. Softw. 53, 1–18 (2013).
van der Walt, S., Colbert, S.C. & Varoquaux, G. The NumPy Array: a structure for efficient numerical computation. Comput. Sci. Eng. 13, 22–30 (2011).
Hunt, M. et al. Circlator: automated circularization of genome assemblies using long sequencing reads. Genome Biol. 16, 294 (2015).
Krumsiek, J., Arnold, R. & Rattei, T. Gepard: a rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics 23, 1026–1028 (2007).
Aziz, R.K. et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9, 75 (2008).
Acknowledgements
We thank M. Lewis for her assistance in DNA extraction and A. Bashir for his guidance in computational matters. We also thank those who contributed to the generation of the publically available SMRT sequencing data for the 20-member Mock Community B. The work is funded by R01 GM114472 (G.F.) from the National Institutes of Health and Icahn Institute for Genomics and Multiscale Biology. G.F. is a Nash Family Research Scholar. This work was also supported in part through the computational resources and staff expertise provided by the Department of Scientific Computing at the Icahn School of Medicine at Mount Sinai.
Author information
Authors and Affiliations
Contributions
J.B. and G.F. designed the methods. J.B. developed the software package for all the proposed computational analyses. J.B., E.W.T., J.J.F. R.S., E.E.S. and G.F. contributed to experimental design. I.M., X.-S.Z., A.D.-R., R.C., E.W.T. and J.J.F. conducted the experiments. G.D. and R.S. designed and conducted sequencing. J.B., S.Z., E.W.T., J.J.F., R.S., E.E.S. and G.F. analyzed the data. J.B. and G.F. wrote the manuscript with inputs and comments from all co-authors. G.F. conceived and supervised the project.
Corresponding author
Ethics declarations
Competing interests
E.E.S. is on the scientific advisory board of Pacific Biosciences. J.B. and G.F. are inventors of a US Provisional patent application (No. 62/525,908) that describes the method for methylation binning.
Integrated supplementary information
Supplementary Figure 1 Binning contigs from 8-species mock community.
(a) t-SNE scatter plot of 5-mer composition profiles for contigs and (b) scatter plot of contig GC-content vs. contig coverage.
Supplementary Figure 2 Shorter contigs contain fewer methylated motif sites.
After de novo assembly of reads from a mixture of eight bacterial species, the contigs belonging to C. bolteae were isolated. As the contig length decreases, it becomes less common for the contig to contain IPD values from the full diversity of motif sites that are methylated in C. bolteae, making it increasingly difficult to segregate smaller contigs based on contig methylation patterns alone.
Supplementary Figure 3 Composition and coverage-based binning methods applied to adult mouse gut microbiome assembly.
(a) Contig GC-content vs. coverage for adult mouse gut microbiome assembly, and (b) contig coverage plotted against the contig coverage using sequencing from a related sample.
Supplementary Figure 4 Infant gut microbiome contigs binned by sequence composition and methylation profiles.
(a) t-SNE map of 5-mer frequency features for contigs assembled from a mixture of two infant microbiome samples. Several clusters contain a mixture of species from the same genus. (b) t-SNE map of methylation features for the same contigs. (c) t-SNE map of the same contigs binned by both 5-mer frequency and methylation profiles (Online Methods), which resolve the contigs into mostly species-specific clusters. Kraken annotation relies on an existing reference database (Online Methods) and is therefore incomplete; contigs not generating a database hit are marked Unlabeled. Contigs <10kb are omitted.
Supplementary Figure 5 CONCOCT bins of the mouse gut microbiome.
Taxonomic composition of the 29 bins identified by CONCOCT in the mouse gut metagenomic assembly. Taxonomy is based on contig-level annotations by Kraken.
Supplementary Figure 6 Heatmaps of methylation profiles for K. pneumoniae.
(a) Hierarchical clustering of all known methylated motifs in REBASE for K. pneumoniae strain 234-12 and nine other species whose chromosomes have smaller sequence distance to the K. pneumoniae strain 234-12 plasmid (horizontal red bars) than its own host chromosome. (b) Hierarchical clustering of all motifs in REBASE for 25 strains of K. pneumoniae. The strains contain 17 unique methylation motifs, including CCAYNNNNNTCC that is observed solely in K. pneumoniae strain 234-12.
Supplementary Figure 7 Sequence composition t-SNE map of modified HMP mock community B.
5-mer frequency-based binning of assembled contigs and raw reads (length>15kb) from the log-abundance HMP mock community. Only the contigs are labeled (raw reads represented underneath contigs by density map) and the sum of assembled bases for each Kraken-annotated species is included in the legend.
Supplementary Figure 8 5-mer frequency-based binning of unaligned reads from the modified HMP mock community B.
(a) Read lengths between 5-10kb, and (b) read lengths between 10-15kb. The shorter read lengths result in more diffuse and overlapping clusters due to the increased variation in 5-mer frequency metrics on these shorter reads.
Supplementary Figure 9 t-SNE map of read-level methylation profiles for two H. pylori strains.
2D map of reads from each of the H. pylori strains, 26695 and J99, analyzed in the multi-strain synthetic mixture. 2D map generated using t-SNE, where the only features used in dimensionality reduction are methylation profiles of the reads.
Supplementary Figure 10 Comparison of abundance-matched SMRT vs. synthetic long read (SLR) sequencing coverage.
(a) Human Microbiome Project Mock Community B members in decreasing order of GC content in genome. The percentage of the reference positions covered by SLRs is consistently lower than the percentage covered by abundance-matched SMRT reads. (b) Coverage variation for alignments of abundance-matched SLR and SMRT reads. A significant number of bases in SLRs are aligned in the same regions, creating dramatic peaks in coverage. SMRT reads largely lack these peaks and have a more uniform coverage profile.
Supplementary Figure 11 Examples of uneven coverage in SLR.
Uneven coverage by synthetic long reads in a 40 kb region of the S. agalactiae genome (a), a 40 kb region of the S. aureus genome (b), and a 50 kb region of the P. aeruginosa genome (c).
Supplementary Figure 12 Genomewide coverage of SLR and SMRT reads for all genomes in HMP mock community B.
Genome-wide coverage of abundance-matched synthetic long reads (red lines) and SMRT reads (blue lines). Regions with zero coverage are highlighted for synthetic long reads (pink) and SMRT reads (light blue).
Supplementary Figure 13 Reference matches for bins identified from methylation profiles in mouse gut microbiome.
Dot plot visualizations created using mummerplot that show the top reference alignment for bins isolated from the mouse gut microbiome metagenomic assembly using only methylation profiles. See Supplementary Table 6 for details of these alignments and the matching reference sequences.
Supplementary Figure 14 Modified relative abundances in HMP mock community B.
Relative abundances of the 20-species in the Human Microbiome Project mock community B modified to follow a log-curve distribution.
Supplementary Figure 15 Sequence composition t-SNE map of unmodified HMP mock community B.
5-mer frequency-based binning of assembled contigs and raw reads (length>15kb) from the even-abundance HMP mock community B. Only the contigs are labeled (raw reads represented underneath contigs by density map) and the sum of assembled bases for each Kraken-annotated species is included in the legend.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–15 Supplementary Methods (PDF 2223 kb)
Supplementary Tables
Supplementary tables 1–11 (ZIP 465 kb)
Supplementary Code
Mbin Software package and relevant scripts (ZIP 43 kb)
Rights and permissions
About this article
Cite this article
Beaulaurier, J., Zhu, S., Deikus, G. et al. Metagenomic binning and association of plasmids with bacterial host genomes using DNA methylation. Nat Biotechnol 36, 61–69 (2018). https://doi.org/10.1038/nbt.4037
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.4037
This article is cited by
-
mEnrich-seq: methylation-guided enrichment sequencing of bacterial taxa of interest from microbiome
Nature Methods (2024)
-
Plasmids, a molecular cornerstone of antimicrobial resistance in the One Health era
Nature Reviews Microbiology (2024)
-
Long-read assembled metagenomic approaches improve our understanding on metabolic potentials of microbial community in mangrove sediments
Microbiome (2023)
-
Navigating the pitfalls of mapping DNA and RNA modifications
Nature Reviews Genetics (2023)
-
A high-quality genome compendium of the human gut microbiome of Inner Mongolians
Nature Microbiology (2023)