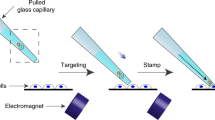
Abstract
Genetic engineering by viral infection of single cells is useful to study complex systems such as the brain. However, available methods for infecting single cells have drawbacks that limit their applications. Here we describe 'virus stamping', in which viruses are reversibly bound to a delivery vehicle—a functionalized glass pipette tip or magnetic nanoparticles in a pipette—that is brought into physical contact with the target cell on a surface or in tissue, using mechanical or magnetic forces. Different single cells in the same tissue can be infected with different viruses and an individual cell can be simultaneously infected with different viruses. We use rabies, lenti, herpes simplex, and adeno-associated viruses to drive expression of fluorescent markers or a calcium indicator in target cells in cell culture, mouse retina, human brain organoid, and the brains of live mice. Virus stamping provides a versatile solution for targeted single-cell infection of diverse cell types, both in vitro and in vivo.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Callaway, E.M. Transneuronal circuit tracing with neurotropic viruses. Curr. Opin. Neurobiol. 18, 617–623 (2008).
Hsu, P.D., Lander, E.S. & Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278 (2014).
Swiech, L. et al. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat. Biotechnol. 33, 102–106 (2015).
Marshel, J.H., Mori, T., Nielsen, K.J. & Callaway, E.M. Targeting single neuronal networks for gene expression and cell labeling in vivo. Neuron 67, 562–574 (2010).
Rancz, E.A. et al. Transfection via whole-cell recording in vivo: bridging single-cell physiology, genetics and connectomics. Nat. Neurosci. 14, 527–532 (2011).
Wertz, A. et al. PRESYNAPTIC NETWORKS. Single-cell-initiated monosynaptic tracing reveals layer-specific cortical network modules. Science 349, 70–74 (2015).
Fishell, G. & Heintz, N. The neuron identity problem: form meets function. Neuron 80, 602–612 (2013).
Siegert, S. et al. Genetic address book for retinal cell types. Nat. Neurosci. 12, 1197–1204 (2009).
Belgard, T.G. et al. A transcriptomic atlas of mouse neocortical layers. Neuron 71, 605–616 (2011).
Bernard, A. et al. Transcriptional architecture of the primate neocortex. Neuron 73, 1083–1099 (2012).
Turner, D.L. & Cepko, C.L. A common progenitor for neurons and glia persists in rat retina late in development. Nature 328, 131–136 (1987).
Walsh, C. & Cepko, C.L. Clonally related cortical cells show several migration patterns. Science 241, 1342–1345 (1988).
Eiraku, M. et al. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 472, 51–56 (2011).
Kadoshima, T. et al. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc. Natl. Acad. Sci. USA 110, 20284–20289 (2013).
Lancaster, M.A. & Knoblich, J.A. Organogenesis in a dish: modeling development and disease using organoid technologies. Science 345, 1247125 (2014).
Vélez-Fort, M. et al. The stimulus selectivity and connectivity of layer six principal cells reveals cortical microcircuits underlying visual processing. Neuron 83, 1431–1443 (2014).
Stiefel, P. et al. Cooperative vaccinia infection demonstrated at the single-cell level using FluidFM. Nano Lett. 12, 4219–4227 (2012).
Martínez-Martín, D. et al. Inertial picobalance reveals fast mass fluctuations in mammalian cells. Nature 550, 500–505 (2017).
Nguyen, T.D. et al. Targeted single-neuron infection with rabies virus for transneuronal multisynaptic tracing. J. Neurosci. Methods 209, 367–370 (2012).
Cronin, J., Zhang, X.-Y. & Reiser, J. Altering the tropism of lentiviral vectors through pseudotyping. Curr. Gene Ther. 5, 387–398 (2005).
Wickersham, I.R., Sullivan, H.A. & Seung, H.S. Axonal and subcellular labelling using modified rabies viral vectors. Nat. Commun. 4, 2332 (2013).
Haberl, M.G. et al. An anterograde rabies virus vector for high-resolution large-scale reconstruction of 3D neuron morphology. Brain Struct. Funct. 220, 1369–1379 (2015).
Mothes, W., Sherer, N.M., Jin, J. & Zhong, P. Virus cell-to-cell transmission. J. Virol. 84, 8360–8368 (2010).
Burns, J.C., Friedmann, T., Driever, W., Burrascano, M. & Yee, J.K. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc. Natl. Acad. Sci. USA 90, 8033–8037 (1993).
Carneiro, F.A., Bianconi, M.L., Weissmüller, G., Stauffer, F. & Da Poian, A.T. Membrane recognition by vesicular stomatitis virus involves enthalpy-driven protein-lipid interactions. J. Virol. 76, 3756–3764 (2002).
Conti, C., Mastromarino, P., Riccioli, A. & Orsi, N. Electrostatic interactions in the early events of VSV infection. Res. Virol. 142, 17–24 (1991).
Bailey, S.N., Ali, S.M., Carpenter, A.E., Higgins, C.O. & Sabatini, D.M. Microarrays of lentiviruses for gene function screens in immortalized and primary cells. Nat. Methods 3, 117–122 (2006).
Lei, P., Bajaj, B. & Andreadis, S.T. Retrovirus-associated heparan sulfate mediates immobilization and gene transfer on recombinant fibronectin. J. Virol. 76, 8722–8728 (2002).
Wegmann, F. et al. Polyethyleneimine is a potent mucosal adjuvant for viral glycoprotein antigens. Nat. Biotechnol. 30, 883–888 (2012).
Nakagawa, T. et al. Fabrication of amino silane-coated microchip for DNA extraction from whole blood. J. Biotechnol. 116, 105–111 (2005).
Hunt, L.A. & Summers, D.F. Glycosylation of vesicular stomatitis virus glycoprotein in virus-infected HeLa cells. J. Virol. 20, 646–657 (1976).
Elaissari, A. Colloidal Biomolecules, Biomaterials, and Biomedical Applications (CRC Press, 2003).
Barde, I., Salmon, P. & Trono, D. Production and titration of lentiviral vectors. 53, 4.21 Curr. Protoc. Neurosci. (2010).
Hatori, M. et al. Inducible ablation of melanopsin-expressing retinal ganglion cells reveals their central role in non-image forming visual responses. PLoS One 3, e2451 (2008).
Judkewitz, B., Rizzi, M., Kitamura, K. & Häusser, M. Targeted single-cell electroporation of mammalian neurons in vivo. Nat. Protoc. 4, 862–869 (2009).
Chen, T.-W. et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013).
Deverman, B.E. et al. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat. Biotechnol. 34, 204–209 (2016).
Hillier, D. et al. Causal evidence for retina-dependent and -independent visual motion computations in mouse cortex. Nat. Neurosci. 20, 960–968 (2017).
Lancaster, M.A. et al. Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379 (2013).
Drokhlyansky, E. et al. The brain parenchyma has a type I interferon response that can limit virus spread. Proc. Natl. Acad. Sci. USA 114, E95–E104 (2017).
Alsteens, D. et al. Nanomechanical mapping of first binding steps of a virus to animal cells. Nat. Nanotechnol. 12, 177–183 (2017).
Muñoz, W., Tremblay, R. & Rudy, B. Channelrhodopsin-assisted patching: in vivo recording of genetically and morphologically identified neurons throughout the brain. Cell Reports 9, 2304–2316 (2014).
Wickersham, I.R., Finke, S., Conzelmann, K.-K. & Callaway, E.M. Retrograde neuronal tracing with a deletion-mutant rabies virus. Nat. Methods 4, 47–49 (2007).
Wickersham, I.R., Sullivan, H.A. & Seung, H.S. Production of glycoprotein-deleted rabies viruses for monosynaptic tracing and high-level gene expression in neurons. Nat. Protoc. 5, 595–606 (2010).
Gomme, E.A., Faul, E.J., Flomenberg, P., McGettigan, J.P. & Schnell, M.J. Characterization of a single-cycle rabies virus-based vaccine vector. J. Virol. 84, 2820–2831 (2010).
Yonehara, K. et al. The first stage of cardinal direction selectivity is localized to the dendrites of retinal ganglion cells. Neuron 79, 1078–1085 (2013).
Madisen, L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010).
Müller, J., Bakkum, D.J. & Hierlemann, A. Sub-millisecond closed-loop feedback stimulation between arbitrary sets of individual neurons. Front. Neural Circuits 6, 121 (2013).
Martinez-Martin, D. et al. Resolving structure and mechanical properties at the nanoscale of viruses with frequency modulation atomic force microscopy. PLoS One 7, e30204 (2012).
Farrow, K. et al. Ambient illumination toggles a neuronal circuit switch in the retina and visual perception at cone threshold. Neuron 78, 325–338 (2013).
Gogolla, N., Galimberti, I., DePaola, V. & Caroni, P. Preparation of organotypic hippocampal slice cultures for long-term live imaging. Nat. Protoc. 1, 1165–1171 (2006).
Lancaster, M.A. & Knoblich, J.A. Generation of cerebral organoids from human pluripotent stem cells. Nat. Protoc. 9, 2329–2340 (2014).
Acknowledgements
We thank J. Gründemann for technical assistance setting up the brain slice procedure, J.M. Mateos, A. Kaech, and J. Doehner from the Zurich Center of Microscopy and Image Analysis (ZMB), U. Schwarz from Leica Mannheim, and T. Horn from the DBSSE imaging facility for helping with imaging and data preparation, M.J. Schnell for providing the BSR-VSV-RVG cell line, E.M. Callaway for providing the B7GG cell line, C.P. Patino Alvarez and A. Villemain for producing viruses, Helbling Technik Bern AG for technical assistance modeling the magnet used for shielded virus stamping, and the members of the Roska laboratory for technical assistance. The study was supported by a European Union grant (FP7/211800) to D.J.M.; a Human Frontier Science Program Long-Term Fellowship (LT000173/2013-L) and a Swiss National Science Foundation Ambizione Fellowship to S.T.; a European Molecular Biology Organization Long-Term Fellowship (506-2012) to D.M.M.; Swiss National Science Foundation grants (310030B_160225 to D.J.M. and 3100330B_163457 to B.R.), the National Center of Competence in Research Molecular Systems Engineering grant to D.J.M. and B.R.; European Research Council (669157, RETMUS), DARPA (HR0011-17-C-0038, Cortical Sight) grants to B.R., a Deutsche Forschungsgemeinschaft grant (SFB870) to K.K.C.
Author information
Authors and Affiliations
Contributions
Experiments were designed by R.S., S.T., K.B., G.K., G.F., D.J.M., and B.R. G-deleted rabies variants were made by A.G. and K.K.C. Cell cultures, immunohistochemistry, electron microscopy, and confocal microscopy were performed by R.S. with the exception of in vivo samples, which were processed by A.W., and organoid samples, which were processed by M.M. Viruses were prepared by R.S., K.B., M.A.M., R.N., and K.Y. Pipettes for unshielded virus stamping were prepared by D.M.M. Brain slice preparations, retinal preparations, unshielded virus stamping, and tissue cultures were performed by S.T. and K.B. The magnetic forces related to the magnet used for shielded stamping were measured and modeled by R.S. and C.S.C. Sequential multi-day single-cell infection experiments were performed by R.S. and C.S.C. Magnetic nanoparticle preparations and shielded stamping in cell culture were performed by R.S. In vivo nanoparticle optimization was performed by D.H. Shielded in vivo stamping was performed by S.T. and G.K. Organoids were grown by M.M., J.K., and B.G.S. Shielded organoid stamping was performed by G.K. and M.M. In vivo two-photon calcium imaging was performed by G.K. Computer reconstructions were performed by R.S., M.A.M., and A.P. Figures were made by R.S., S.T., M.M., and G.K. The paper was written by R.S., S.T., D.J.M., and B.R.
Corresponding authors
Ethics declarations
Competing interests
R.S., S.T., G.F., D.J.M. and B.R. applied for a patent related to viral stamping approach.
Integrated supplementary information
Supplementary Figure 1 Preparing glass pipettes for unshielded virus stamping of surface cells and performing virus stamping.
The tools used for the four steps described in Methods (‘Glass pipette preparation’) are shown: a, Pulling and blunting of pipettes; b, Cleaning of pipettes; c, Silane functionalization of pipettes; d, Virus binding to pipettes. e, Non-targeted infection can be averted by using a lower virus concentration. The percentage of preparations in which non-targeted infection was observed when binding different concentrations of VSVG-coated rabies viruses encoding GFP (indicated on the x-axis) to functionalized glass pipettes (for each virus concentration, n = 8). For these experiments, virus-bound pipette tips were lowered into, and then removed from, solution containing plated BHK cells. The graph indicates that diluting the virus 100-fold eliminated non-targeted infection completely. The starting titer of the virus was 109 plaque-forming units per ml. f, Infrared image of a flame-blunted, virus-bound glass patch pipette that is touching the cell body of a retinal ganglion cell during virus stamping. Scale bar, 20 μm. g, A single BHK cell was targeted with a pipette tip bound with VSVG-coated rabies viruses encoding tdTomato. Time series of superimposed phase contrast and fluorescence images, showing the development of fluorescence in a single BHK cells over a 24 hours period. The insets in the bottom images are higher-resolution views of the infected cell. The black lines on the top and right are the walls of the well in which the cells were cultured. Scale bars, 200 μm, and 10 μm for inset.
Supplementary Figure 2 Versatility of virus stamping.
a,b, Targeted single cell infection by combining virus stamping with 2-photon microscopy. A neuronal cell culture was transfected with a low concentration of plasmids encoding GFP, resulting in sparse GFP labeling (left). A single GFP-expressing neuron was then targeted with VSVG-coated rabies viruses encoding tdTomato, leading to a single neuron expressing both tdTomato (middle image) and GFP (right image). Scale bar, 50 μm. c-f, Co-infection of single cells by virus stamping using one pipette bound with two different viruses. VSVG-coated rabies viruses encoding tdTomato and VSVG-coated lenti viruses encoding GFP were used. Examples of single cell co-infection in neuron culture (c-d, scale bar, 20 μm), and brain slice (e-f, scale bar, 60 μm). g-h, Monosynaptic circuit tracing in brain slice initiated by co-infecting a single cell with VSVG-coated G-deleted rabies viruses encoding GFP and VSVG-coated lenti viruses encoding the rabies glycoprotein (Rabies-G). The target cell (left image, white arrow) was confirmed by staining against the rabies glycoprotein, which only labeled a single cell in the slice. Scale bar, 50 μm. i, Multi-day, sequential single cell virus stamping. On day 1 at t = 0 hours, a single cortical neuron was targeted with a virus stamper bound with VSVG-coated G-deleted rabies viruses encoding GFP. j, After 48 hours, the infected cell expressed GFP and it was then stamped a second time with a virus stamper bound with HSVs encoding mCherry. k, Four hours later the stamped cell expressed both GFP and mCherry. Note that the stamped cell migrated slightly in the culture dish and changed shape slightly over the course of the experiment. The stamped cell was tracked over time to ensure that no neighboring cells showed fluorescent protein expression. Scale bar, 20 μm.
Supplementary Figure 3 Demonstration that virus stamping works with non-VSVG coated enveloped viruses and non-enveloped viruses, and that virus stamped neurons survive for many days post-infection.
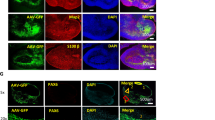
a, A single cultured cortical neuron (embryonic day 18, from rat) was stamped shielded virus stamper that contained magnetic nanoparticles coated with both herpes simplex viruses (HSV1, non-VSVG coated) encoding GFP and G-coated rabies viruses encoding tdTomato. Both fluorophores expressed for a week and the neuron kept its morphology throughout this period. Scale bar, 50 μm. Note that adult neurons cannot be stamped at their soma with G-coated rabies or HSV1 viruses since these viruses enter adult neurons at their axon terminals. b, A single cultured cortical neuron was simultaneously stamped with two different AAVs (non-enveloped virus), one encoding GCaMP6 s and the other encoding mRuby. After 2 weeks, both fluorophores strongly expressed and the neuron maintained a healthy morphological appearance. Note than in general, AAVs take longer to express their payload than the other virus types used in this study. Scale bar, 20 μm.
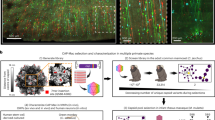
Supplementary Figure 4 Cell infection with virus-coated magnetic nanoparticles.
a, For these experiments, nanoparticle-containing virus solution was applied to cell culture and a permanent magnet was used to pull the nanoparticles into contact with the cells. Confocal image of HEK cells infected with AEEA-functionalized nanoparticles (Alpha Biobeads, see Methods) bound with VSVG-coated rabies viruses encoding tdTomato. b, Confocal image of HEK cells infected with cationically-functionalized nanoparticles (OZ Bioscience, see Methods) bound with VSVG-coated rabies viruses encoding tdTomato. Scale bars, 50 μm. c, Quantification of cells infected in a 200 μm2 field of view in the center of a 24 well plate for both cationically and AEEA-functionalized nanoparticles bound with VSVG-coated rabies viruses encoding tdTomato. Red lines represent mean values. d, Testing the efficiency of virus-bound magnetic nanoparticles for stamping. Alexa 488-coated magnetic nanoparticles were bound with VSVG-coated lenti viruses encoding tdTomato. Single, highly fluorescent nanoparticles were FACS-isolated and were dropped into the wells of a glass bottom 96 well plate that had been plated with a confluent monolayer of HeLa cells (see Methods). A permanent magnet was used to pull the nanoparticles into contact with the cells. e, Example infection in one of the wells showing low resolution (top) and magnified (bottom) DIC and fluorescence images of a single infected cell and the fluorescent nanoparticle. Scale bars, 100 μm (top) and 50 μm (bottom). f, Percentage of wells in a 96 well plate that contained a fluorescent cell. g, Nanoparticles bind viruses with high efficiency. A 10 μl solution of VSVG-coated lenti viruses encoding GFP was mixed with 10 μl magnetic nanoparticle solution in a 1.5 ml centrifuge tube. The nanoparticles were pulled down to the bottom of the tube with a permanent magnet (see Methods). The supernatant was isolated and used for testing the infectivity of free viruses that were not bound to nanoparticles. The nanoparticles were then diluted with 10 μl of L-15 media and the magnet was removed. This nanoparticle-containing virus solution was then used to test the infectivity by placing 1 μl of nanoparticle solution per well of a 24 well plate and using a permanent magnet to pull the nanoparticles into contact with the cells. Top row, confocal images showing cultured HEK cells infected with nanoparticle-containing fraction. Bottom row, cultured HEK cells infected with the supernatant. Left column, DAPI staining. Middle column, GFP fluorescence. Right column, overlay of DAPI and GFP signals. Scale bar, 20 μm. h, Number of cells infected in a 200 μm2 field of view in the center of a 24 well plate for nanoparticle-containing fraction and supernatant. Red lines represent mean values. i, Virus stamping of single cells in cell culture with a shielded virus stamper. Targeted infection of one of the two GFP positive cells with VSVG-coated rabies viruses encoding tdTomato is shown 24 hours after infection. Left panel, GFP fluorescence. Middle panel, tdTomato fluorescence. Right panel, overlay of GFP and tdTomato signals. Scale bar, 20 μm.
Supplementary Figure 5 Calcium imaging in mouse visual cortex.
a, A schematic of the experimental protocol, in which a mouse with a cranial window above primary visual cortex is placed under a 2-photon microscope, and GCaMP6s based calcium signals are monitored while the lightly anesthetized animal is presented with visual stimulation consisting of 8 directions of drifting grating visual stimulation. b, The shielded virus stamper was used to deliver VSVG-coated G-deleted rabies viruses encoding GCaMP6s to visual cortex. Robust calcium responses were present six days later. The light responses from two cells are shown. Black traces indicate the average from all six trials. c, The PHP.B serotype of AAV was systemically delivered to drive wide-scale infection with GCaMP6s. A mouse was placed under a 2-photon microscope and calcium responses were measured during visual stimulation before and immediately after the electromagnet used for shielded stamping experiments was turned on for ten minutes. The top image shows the field of view of the imaging session and four cells of interest are indicated in red. The light responses of these cells are shown on the bottom in red. The middle image shows the same field of view except after the magnet was turned on for ten minutes. The same four cells on interest are indicated in blue, and their light responses are shown on the bottom. The red and blue calcium traces on the bottom represent averages from six trials. d, The PHP.B serotype of AAV was systemically delivered to drive wide-scale infection with GCaMP6s. Subsequently, magnetic nanoparticles were injected into visual cortex. A week later, the mouse was placed under a 2-photon microscope and calcium responses were measured during visual stimulation. The top image shows the field of view with five cells of interest indicated. The bottom traces show the light responses to the five cells of interest. The responses represent the average of six trials.
Supplementary Figure 6 Estimating the guidance of magnetic nanoparticles within the shielded stamping pipette by the externally applied magnetic field.
a, Schematic setup used for shielded virus stamping for in vivo experiments. The highlighted area of the electromagnet illustrates the 2D axisymmetric space used to simplify the finite element modeling of the magnetic field created by the magnet (Supplementary Note 1). Variables that are critical for stamping that adjust the magnetic field are 1) the current driving the electromagnet, 2) the distance between the electromagnet and the stamping pipette (indicated by ‘d’ in the figure), and 3) the angle between electromagnet and pipette (indicated at 45° in the figure). b, Experimentally determined (mean ± 3 standard errors) and model-predicted B-field values, which falls exponentially with increasing distance between the electromagnet and the pipette (coefficient of determination = 0.984). c, Map of the force exerted on a 100 nm sized (diameter) magnetic nanoparticle in the vicinity of the electromagnet. White lines indicate the direction of the force-field. Scale bar, 1 cm. d, Nanoparticles attached to the surface of a Hela cell. Nanoparticles (Alpha Biosciences) conjugated to both a fluorophore (red) and VSVG-coated lenti viruses encoding GFP (green) were pulled from a stamping pipette by the electromagnet. Scale bar, 10 μm.
Supplementary Figure 7 Magnetic nanoparticles concentrate viruses and drive robust infection.
a, Confocal images of HEK cells co-infected with magnetic nanoparticles bound with VSVG-coated G-deleted rabies viruses, G-coated G-deleted rabies viruses, AAVs and HSVs. For this experiment, nanoparticle-containing virus solution was applied to the cell culture and a permanent magnet was used to pull the nanoparticles into contact with the cells. Scale bar, 20 μm. b, Confocal images showing HEK cells co-infected with free VSVG-coated rabies, G-coated rabies, HSVs and AAVs that are not bound to nanoparticles (i.e. the virus solution was directly applied to the cells; bottom image). Scale bar, 20 μm. Note that combining the viruses with the nanoparticles and guiding the nanoparticles towards the cells with a magnet greatly increases infectivity. c, The number of single cells co-infected with all four viruses applied in the experiments with virus-bound nanoparticles or viruses alone. The red lines represent mean. d, Infectivity of AAV virus-bound nanoparticles. Serotype 2/1 AAVs encoding tdTomato were bound to magnetic nanoparticles in a centrifuge tube. The virus-bound nanoparticles were pulled to the bottom of a centrifuge tube using a magnet and were injected locally in layer 5 of mouse cortex. The supernatant was separated and was also injected nearby into the cortex.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 (PDF 1678 kb)
Supplementary Note 1
Modeling the electromagnet used for virus stamping and its interaction with the virus-coated nanoparticles (PDF 524 kb)
Rights and permissions
About this article
Cite this article
Schubert, R., Trenholm, S., Balint, K. et al. Virus stamping for targeted single-cell infection in vitro and in vivo. Nat Biotechnol 36, 81–88 (2018). https://doi.org/10.1038/nbt.4034
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.4034
This article is cited by
-
CRISPR-clear imaging of melanin-rich B16-derived solid tumors
Communications Biology (2023)
-
AAV capsid variants with brain-wide transgene expression and decreased liver targeting after intravenous delivery in mouse and marmoset
Nature Neuroscience (2022)
-
Accurate signal-source localization in brain slices by means of high-density microelectrode arrays
Scientific Reports (2019)
-
Next-generation interfaces for studying neural function
Nature Biotechnology (2019)
-
Magnetically guided virus stamping for the targeted infection of single cells or groups of cells
Nature Protocols (2019)